|
 |
 |
 |
 |
 |
- Overview
- Available TEPs
- The TEP Target List
- How do I nominate targets as possible TEPs?
- How we generate and disseminate TEPs
Overview

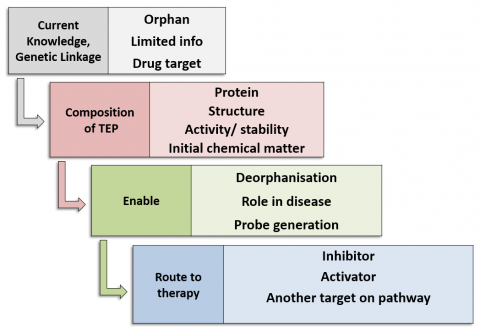
The Target Enabling Package (TEP) programme's foundation is built upon the recognition that genetic data is proving to be a powerful tool for target validation. As such, TEPs provide a critical mass of reagents and knowledge on a protein target to allow rapid biochemical and chemical exploration and characterisation of proteins with genetic linkage to key disease areas. TEPs provide an answer to the missing link between genomics and chemical biology, provide a starting point for chemical probe generation and therefore catalyse new biology and disease understanding with the ultimate aim of enabling translation collaborations and target/ drug discovery.
We are committed to generating and making available 27 high-quality TEPs by June 2020.
For more information regarding any aspect of TEPs and the TEP programme, please email at teps@thesgc.org.
Each TEP, as a minimum, contains:
- Protein production methods
- Biochemical/biophysical assays for activity, affinity
- Structures of the protein, potentially including wild type and disease mutant proteins; full-length or domains; protein-ligand complexes; structures of close homologues
- Initial chemical matter from a fragment or small molecule screen
Additional components of TEPs may be developed on a case-by-case basis, based upon reasonable scientific need, in collaboration with TEP target nominators:
- An antibody or nanobody
- Cell-based assay
- CRISPR knockout
Available TEPs
TEPs that have been approved by our external Evaluation Group are available via the links below.
| Gene | Description | Therapeutic Area | Version | Date |
|---|---|---|---|---|
| SLC12A4/SLC12A6 | Potassium/Chloride Co-transporter 1 and 3 (KCC1/KCC3; SLC12A4/SLC12A6) | Sickle cell disease (SCD), Neurological | Version 1 | 2020 |
| EPB41L3 | EPB41L3 | Neurodegeneration | Version 1 | 2020 |
| INPP5D | INPP5D (SHIP1) | Neurological Disorders | Version 1 | 2020 |
| Moesin | MSN (Moesin) | Alzheimer’s disease | Version 1 | 2020 |
| TNC | Fibrinogen-like globe domain of human Tenascin-C (hFBG-C) | Inflammatory diseases | Version 1 | 2020 |
| ELOVL7 | Elongation of very long chain fatty acids protein 7 (ELOVL7) | Metabolic diseases | Version 1 | 2020 |
| DHTKD1 | Dehydrogenase E1 and transketolase domain-containing protein 1 (DHTKD1) | Metabolic diseases | Version 1 | 2020 |
| DCLRE1C |
Artemis (DCLRE1C, SNM1C) |
Oncology | Version 1 | 2020 |
| TBXT |
Human T-box transcription factor T (Brachyury) |
Cancer | Version 1 | 2020 |
| STAG1 |
Human Stromal Antigen 1 (STAG1) Cohesin Subunit SA-1 |
Cancer | Version 1 | 2020 |
| MTHFR |
Methylene-Tetrahydrofolate Reductase (MTHFR) |
Metabolic disorders, Cancer | Version 2 | 2019 |
| ALAS2 |
Human 5'-Aminolevulinate synthase, erythoid-specific (ALAS2) |
Metabolic diseases | Version 1 | 2019 |
| GALT/GALK1 |
Human Galactose- 1-phosphate uridylyltransferase (GALT) |
Metabolic diseases | Version 1 | 2019 |
| KALRN/RAC1 | Human Kalrin/RAC1 GEF/GTPase Complex (KARLN/RAC1) | Neurological Disorders | Version 1 |
2019 |
| KEAP1 |
Human Kelch-like ECH Associated Protein 1 (KEAP1) |
Neuropsychiatry | Version 1 | 2019 |
| KLHL20 |
Human Kelch-like protein 20 (KLHL20) |
Cancer, Neuropsychiatry | Version 1 | 2019 |
| MLLT1 | Human Mixed-Lineage Leukemia, Translocated to 1 (MLLT1) | Cancer | Version 1 | 2018 |
| TASK1 |
Human TWIK-Related Acid-Sensitive K+ Channel 1 (TASK1) |
Neurological Disorders | Version 2 | 2019 |
| TMEM16K |
Human TMEM16K (ANO10) |
Neurological Disorders | Version 1 | 2019 |
| HCN4 | Human Hyperpolarization Activated Cyclic Nucleotide Gated Ion Channel 4 (HCN4) | Cardiovascular, Inflammation, and Neuro | Version 2 | 2018 |
| NUDT7 | Human Peroxisomal Coenzyme A Diphosphatase NUDT7 (NUDT7) | Metabolic diseases | Version 2 | 2018 |
| PARP14 | Human Poly (ADP-ribose) Polymerase Family Member 14 (PARP14) | Cancer, Inflammation | Version 1 | 2018 |
| RIPK2 | Human Receptor-Interacting Serine/Threonine-Protein Kinase 2 (RIPK2) | Inflammatory diseases | Version 2 | 2018 |
| HAO1 | Human Hydroxyacid Oxidase (HAO1) | Metabolic disorders | Version 2 | 2018 |
| FAM83B | Human Family With Sequence Similarity 83 Member B (FAM83B) | Cancer | Version 1 | 2018 |
| LIMK1 | Human LIM Domain Kinase 1 (LIMK1), Kinase Domain | Neuropsychiatry | Version 6 | 2017 |
| DCLRE1A | Human DNA Cross-Link Repair 1A (DCLRE1A, SNM1A) | Cancer | Version 5 | 2017 |
| DPAGT1 | Human Dolichyl-Phosphate Alpha-N-Acetyl glucosaminyl transferase (DPAGT1) | Neuropsychiatry and neuro genetic disorders | Version 4 | 2017 |
| AASS | Human Alpha-Aminoadipic Semialdehyde Synthase (AASS) | Metabolic & Neurological disorders | Version 6 | 2017 |
| WNK3 | Human With No Lysine Kinase 3 (WNK3) | Metabolic diseases | Version 6 | 2017 |
| HDAC6 | Human Histone Deacetylase 6 (HDAC6) | Oncology | Version 5 | 2016 |
| KDM3B | Lysine Demethylase JMJD1B (KDM3B) | Cancer | Version 5 | 2016 |
| KDM4D | Lysine Demethylase JMJD2D (KDM4D) | Cancer | Version 5 | 2016 |
| PHIP | Human Pleckstrin Homology Domain Interacting Protein (PHIP) | Cancer | Version 5 | 2016 |
| PfBDP4 | Plasmodium bormodomain PfBDP4 | Malaria | Version 5 | 2016 |
| RECQL5 | Human RECQL5 helicase | Cancer | Version 5 | 2016 |
| SETDB1 | Human SET domain bifurcated 1 (SETDB1), Tudor domain | Oncology | Version 6 | 2016 |
| CDK12 | Human Cyclin-Dependent Kinase 12 (CDK12), Kinase Domain | Oncology | Version 7 | 2016 |
The TEP Target List
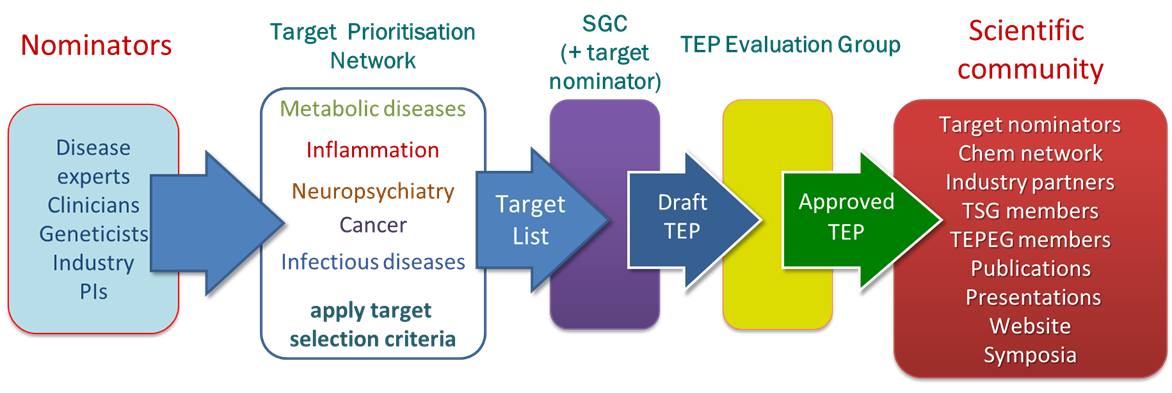
The target list has been derived from nominations from both academia and industry with the expertise in key disease areas that are the focus of this programme: cancer, neuropsychiatry, inflammation, infectious diseases, metabolic disorders and rare diseases. Nominations are prioritised using the following criteria:
- Robustness of available gene-disease association data
- Significance of disease phenotype
- Biological plausibility
- Need for reagents to enable post-gene work (lack of structural and biochemical data)
- Feasibility of producing a high quality TEP with our methodologies
- Prospects for exploiting TEPs, such as existence of downstream communities or collaborators who are committed to using the TEP
Note that becuase the degree of genetic linkage with disease covers a broad spectrum ranging from monogenic (e.g. rare diseases, cancer) through to weaker polygenic (e.g. in some neurological diseases), we do not necessarily prioritise TEP targets based upon such linkages alone. Indeed, we may prioritise targets where there is weak linkage on the basis that TEP components would help to accelerate the understanding of the relevance of that linkage.
The current target list can be downloaded here.
How do I nominate targets as possible TEPs?
The SGC aims to promote exploration of new drug targets for treatment of human diseases in four broad therapeutic areas: Neuropsychiatry, Metabolic and other Rare diseases, Cancer, and Inflammation.
We are seeking nominations of new disease-linked genes, identified through genomic or gene expression research. We will then, possibly in collaboration with the nominating scientist, generate the purified proteins encoded by the disease-linked genes; functional assays; atomic-resolution structures; and chemical starting points. The data and reagents will be available to the wider research community, and in particular to our existing network of collaborators ranging from medicinal chemists to clinicians, which will ensure a rapid impact.
Targets will be prioritised according to the following criteria:
- Robust disease link (Genetic or expression data)
- Plausible therapeutic hypothesis
- Need for tools (proteins, structures, assays, small molecules, antibodies)
- Prospects for collaborative research and downstream utilisation.
To nominate targets please download the Target nomination form and upload using the links below:
![]() Target nomination form (.xlsx)
Target nomination form (.xlsx)
- Neuropsychiatry neurodegeneration:
 Contact
Contact  Nomination form
Nomination form - Metabolic and other rare diseases:
 Contact
Contact  Nomination form
Nomination form - Cancer:
 Contact
Contact  Nomination form
Nomination form - Immune:
 Contact
Contact  Nomination form (See related ULTRA-DD project)
Nomination form (See related ULTRA-DD project)
How we generate and disseminate TEPs
We work closely with disease experts, clinicians, geneticists and industry experts in each of the key disease areas via our Target Prioritisation Networks (TPNs). These networks ensure that we prioritise our work on the targets which are most likely to lead to downstream utility once a TEP is released. The generation of the content of each TEP often directly involves collaborations with the nominator ensuring that that the TEP contains relevant and enabling reagents, data and knowledge.
Each TEP is carefully scrutinised by the TEP Evaluation Group, chaired by Prof. Mike Ferguson (Dundee), which makes the final decision on whether a TEP is suitable to be accepted for release to the public.
Our extensive network of partners and collaborators ensures that our TEPs are rapidly taken up and their contents used to study the biochemistry, structural and chemical biology of the target concerned.