 |
 |
 |
 |
 |
 |
Integral Membrane Proteins
Integral membrane proteins (IMPs) act as the gateways to cells. All cells and organelles are encased in an impermeable lipid bilayer and the IMPs we study are embedded in these membranes. They are the entry and exit routes for many ions, nutrients, waste products, hormones, drugs and large molecules such as proteins and DNA. They are also responsible for much of the communication between cells and their environment. Cells can make a huge variety of these proteins, approximately 30% of the genes in the human genome code for membrane proteins, and yet we know relatively little about these molecules.
Membrane proteins are unfortunately notoriously difficult to handle and study since they are designed to sit within the hydrophobic environment of the lipid bilayer. They tend to be unstable when extracted from their native environment and we have to add detergents to cover the hydrophobic surface. In order to understand and control the function of these proteins it is essential to have information on their three-dimensional structure, which is usually obtained by X-ray crystallography. However the difficulty of handling these proteins has made it difficult to solve the structures, and to date there are fewer than 300 structures of membrane proteins known, less than 0.5 % of all the known structures. For higher eukaryotes the story is even more stark, with only 20 structures of human IMPs and less than 50 mammalian IMPs solved. Membrane proteins are therefore one of the most important remaining frontiers of structural biology research. At the SGC we are now applying our state-of-the-art high-throughput methods to overcome the bottlenecks in membrane protein research, so that we can reliably deliver pure membrane protein samples and structures of these fascinating and medically critical molecules.
Medical importance
The medical importance of this enormous family of proteins cannot be overestimated. Mutations in membrane proteins are involved in many common diseases, including heart disease, where malfunctioning ion channels are often implicated. Drugs targeted to calcium channels can control issues such as high blood pressure and angina. Membrane proteins are involved in cancer, where errors in signalling pathways can lead to cells dividing out of control. Often specific membrane proteins are overproduced in cancer cells and are therefore targets for drug therapy. Diseases of the brain such as migraine, depression and Alzheimer's are all linked to problems with transporters and channels. Cystic fibrosis is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene which encodes a chloride ion channel.
Since many membrane proteins sit at the surface of cells, they are readily available to small molecule drugs circulating in the blood. It is therefore not surprising that over 50% of small molecule drugs bind to membrane proteins. G-protein coupled receptors and channels are particularly important in this respect, but ABC transporters and solute carriers are also targets for drug therapy. Our understanding of many other diseases, and our ability to treat these diseases would benefit greatly from more structural and functional information on the proteins involved. We hope that by solving the structures of these proteins, understanding the underlying biochemistry and interactions with substrates and inhibitors, we can provide more effective treatments for many diseases.
The SGC and integral membrane proteins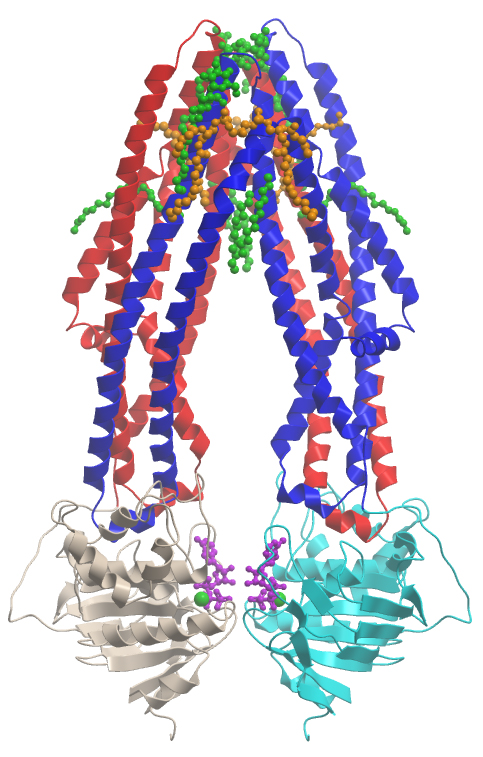
The SGC has extensive experience in solving structures of soluble human proteins using highly efficient, high-throughput systems. In the last two years we have adapted these methods to solving structures for human membrane proteins. We have screened 186 human membrane proteins from diverse families, selecting a series of constructs for each protein, identifying suitable detergents for purification and then scaling up the purificaiton and crystallising these proteins. We have crystallised 3 membrane proteins and solved our first structure, a process which took less than two years from first clone to structure.
We have solved the first structure of a human ABC transporter, a class of proteins which are involved in small molecule transport, multi-drug resistance and diseases such as cystic fibrosis and diabetes. Our first successful target is the human mitochondrial ABC transporter, ABCB10, which is embedded in the inner membrane of mitochondria. ABCB10 is overexpressed during erythroid differentiation, the process which forms red blood cells. it is overexpressed in bone marrow, heart and the liver. There is now evidence that when ABCB10 expression is reduced in cells, they are more susceptible to oxidative stress and that ABCB10 may be involved in protecting the heart during a heart attack.
Technologies for studying IMPs
We are developing generic methods that enable the high throughput determination of structures of human membrane proteins. We selected the baculovirus/insect cell expression system which provides a lipid composition close to that of human cells and is a proven high throughput platform. For each target protein we generate a series of constructs of varying length and different affinity tags, including the full length gene and a series of truncations to remove potentially disordered regions. A high throughput expression screen is used to identify proteins that can be used to purify milligram quantities of IMPS for crystallisation. Each protein is initially purified in dodecyl maltoside (DDM) detergent and is subsequently screened for stability in a series of different detergents to identify the optimum conditions for stability and crystallisation.
We have established methods for high throughput nanodrop crystallisation and manipulation of fragile membrane protein crystals. We have developed efficient systems for screening membrane protein crystals for diffraction quality as a method for crystallisation conditon optimization. We have also optimized our data collection and analysis of diffraction data using intense synchrotron microfocus beamlines, available at resources such as Diamond Light Source Ltd, in Oxfordshire. Wherever possible crystallisation is performed with bound ligands and inhibitors to capture a single native conformation and provide key insights into function and drug design. We also plan to generate antibody fragments against our purified proteins for use as affinity reagents and crystallisation aids.
