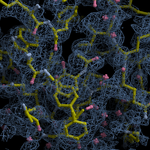
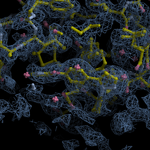
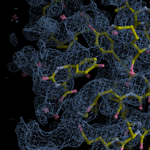
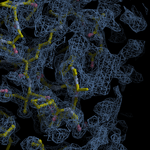
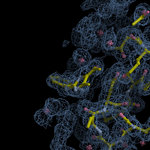
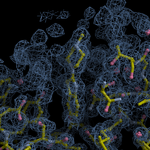
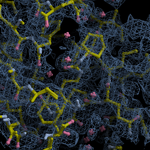
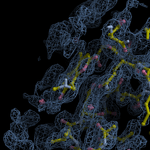
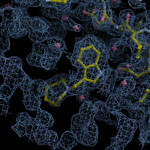
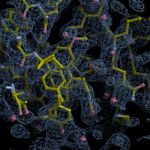
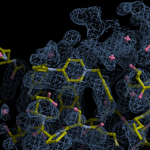
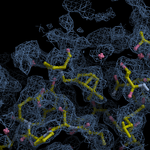
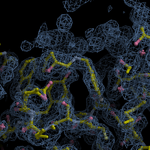
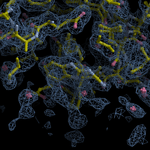
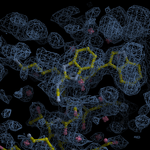
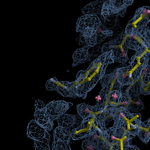
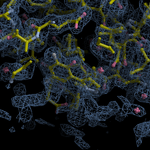
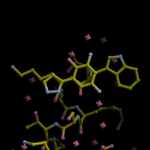
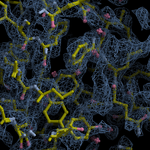
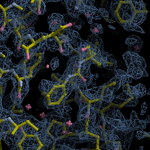
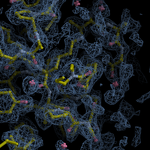
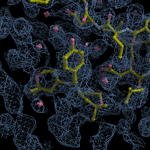
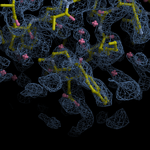
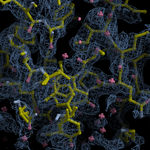
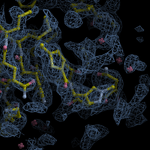
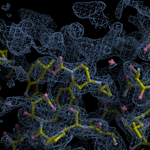
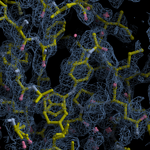
Human DCLRE1AA - XChem results
Click event map in table to view a different crystal / compound pair
Ligand-bound models for Summary
Interpreting 'Ligand confidence'
4 - High Confidence: The expected ligand was easily interpretable from clear density, and subsequent refinement was well-behaved. This ligand can be trusted.
3 - Clear density, unexpected ligand: Density very clearly showed a well-defined ligand, but that ligand was unexpected in that crystal/dataset. The observed ligand was modelled anyway, because its presence could be explained in some way.
2 - Correct ligand, weak density: Though density was weak, it was possible to model the expected ligand, possibly including other circumstantial evidence (e.g. similar ligand in another model).
1 - Low Confidence: The ligand model is to be treated with scepticism, because the evidence (density, identity, pose) were not convincing.
Interpreting 'Model status':
6 - Deposited: The model has been deposited in the PDB.
5 - Deposition ready: The model is fully error-free, in every residue, and is ready for deposition.
4 - CompChem ready: The model is complete and correct in the region of the bound ligand. There may be remaining small errors elsewhere in the structure, but they are far away and unlikely to be relevant to any computational analysis or compound design.
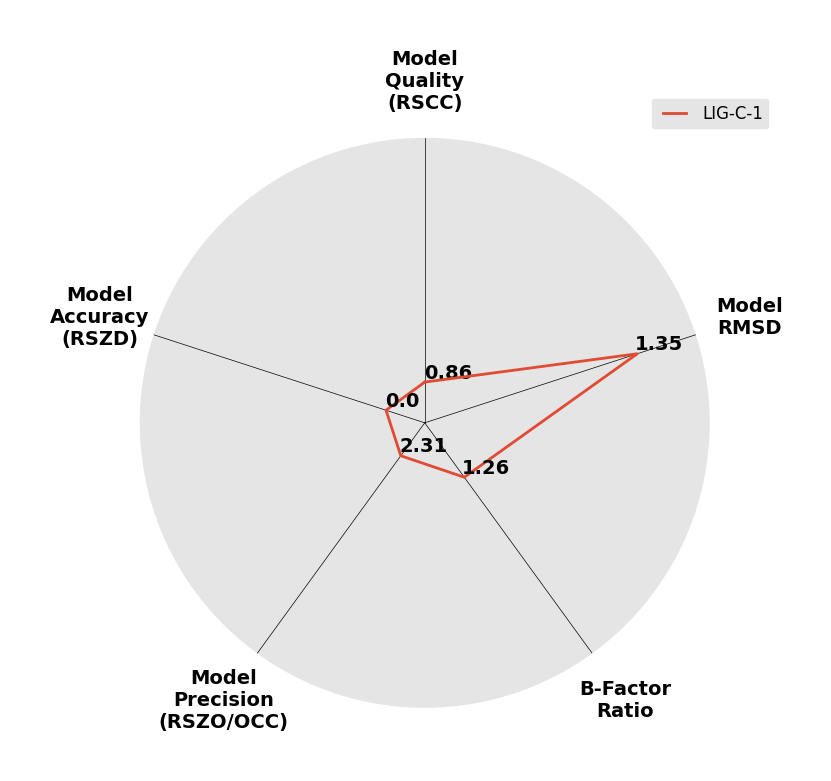
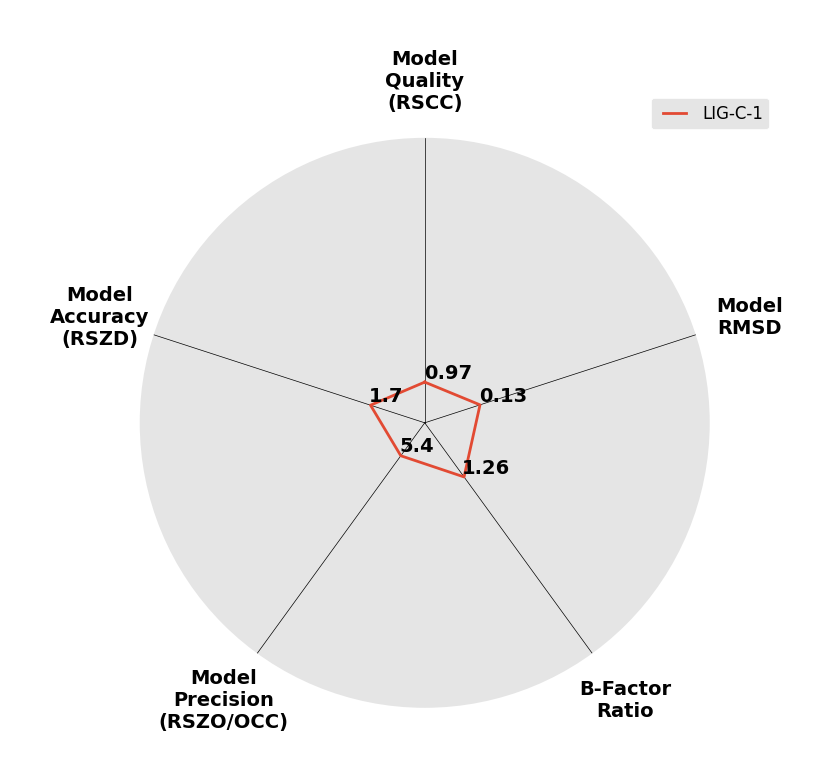
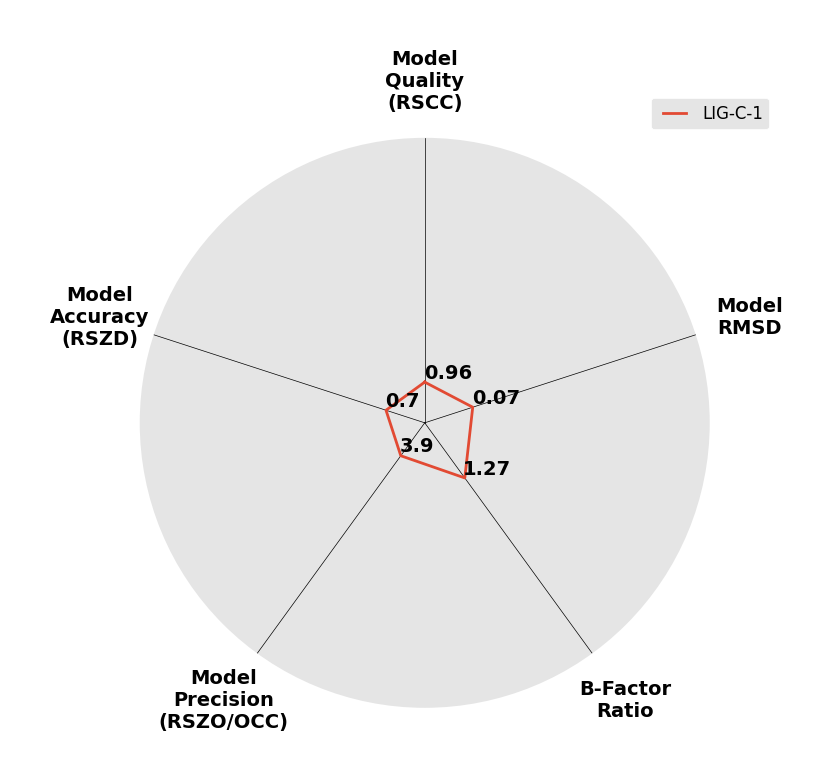
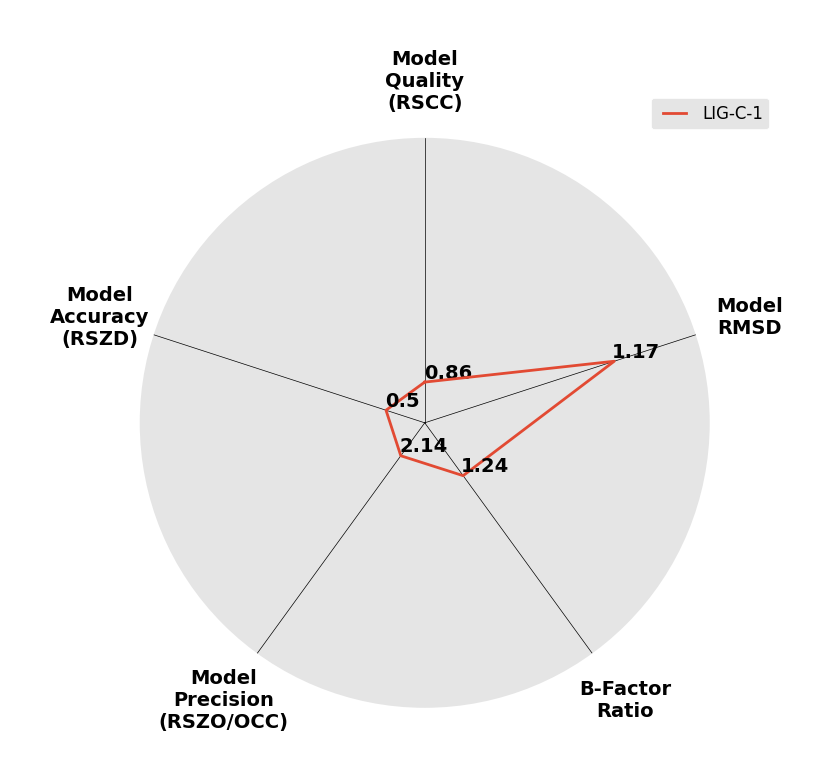
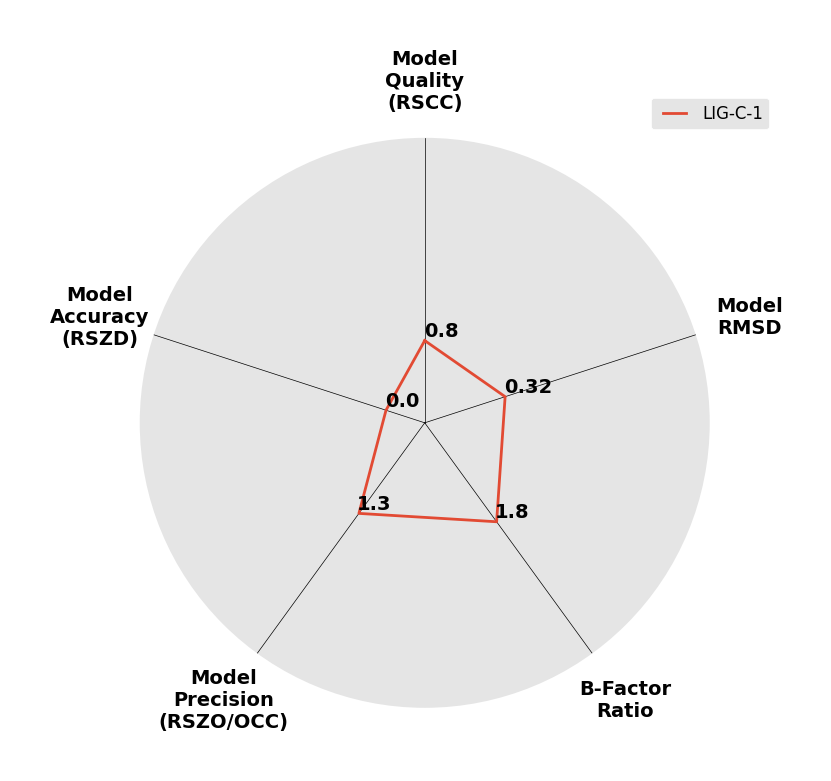
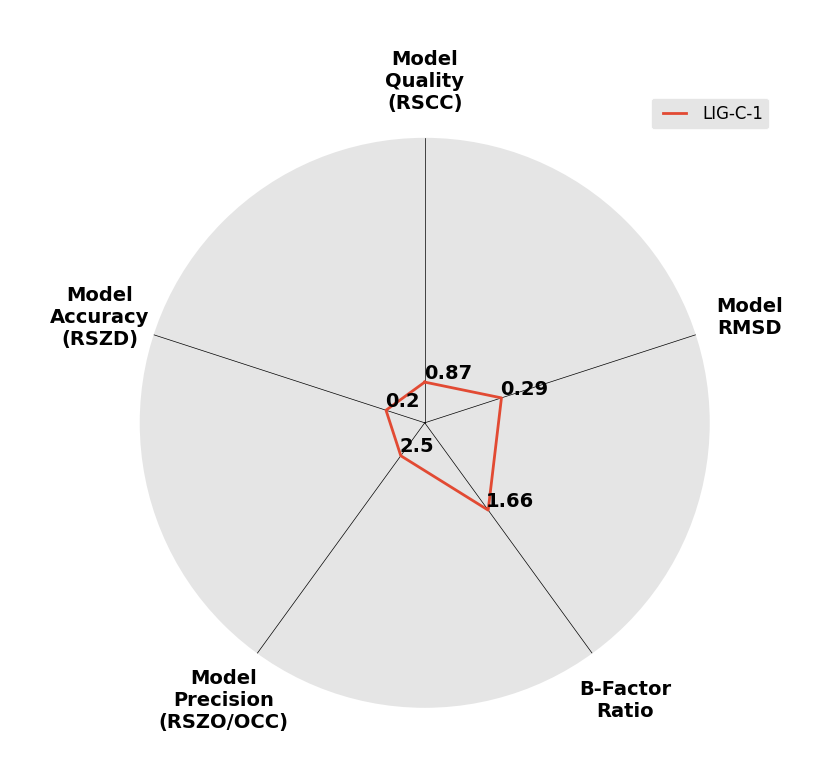
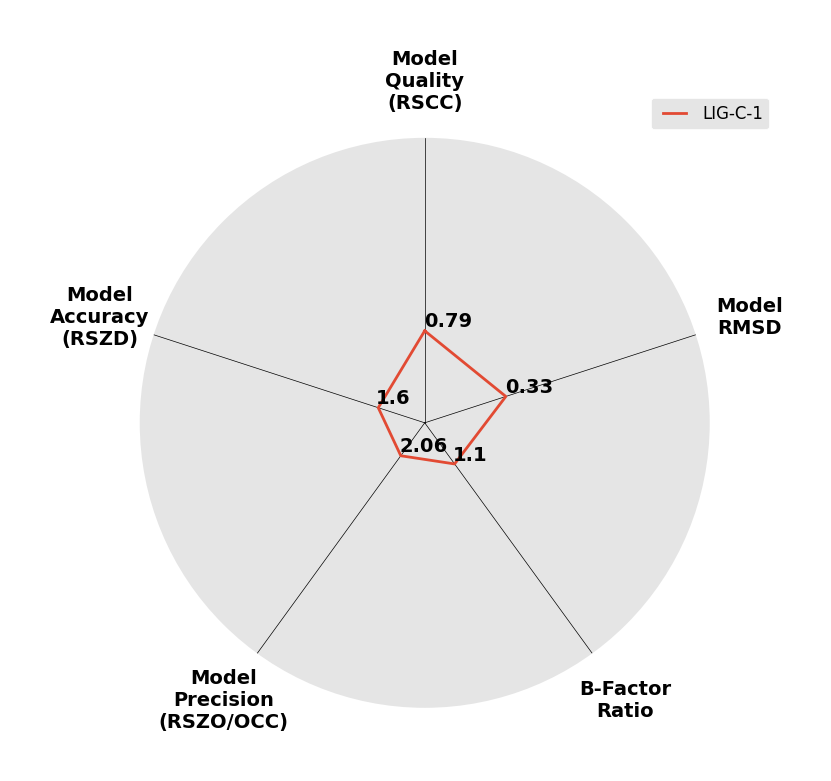
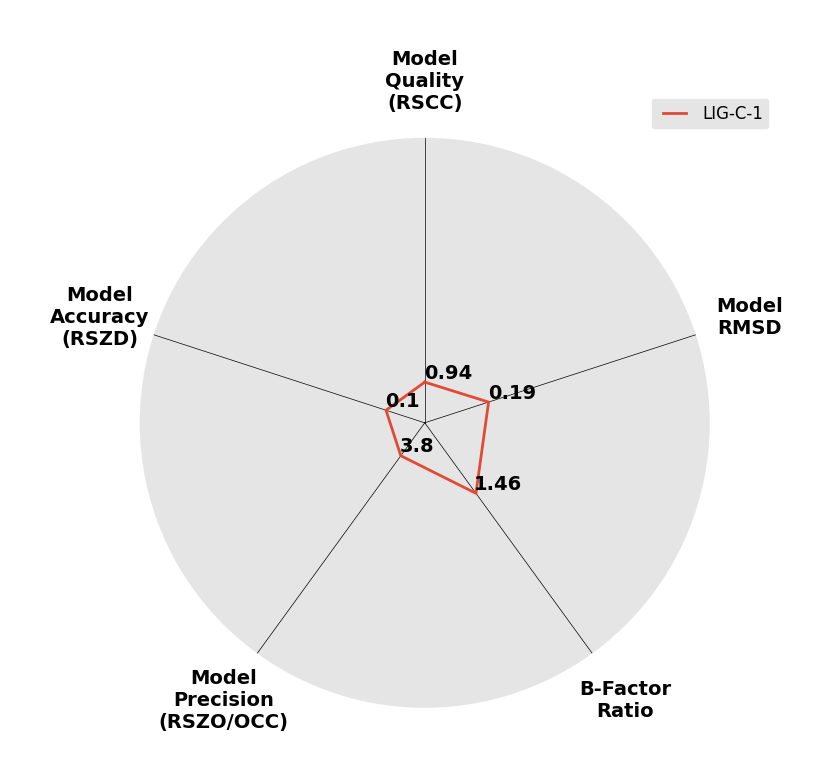
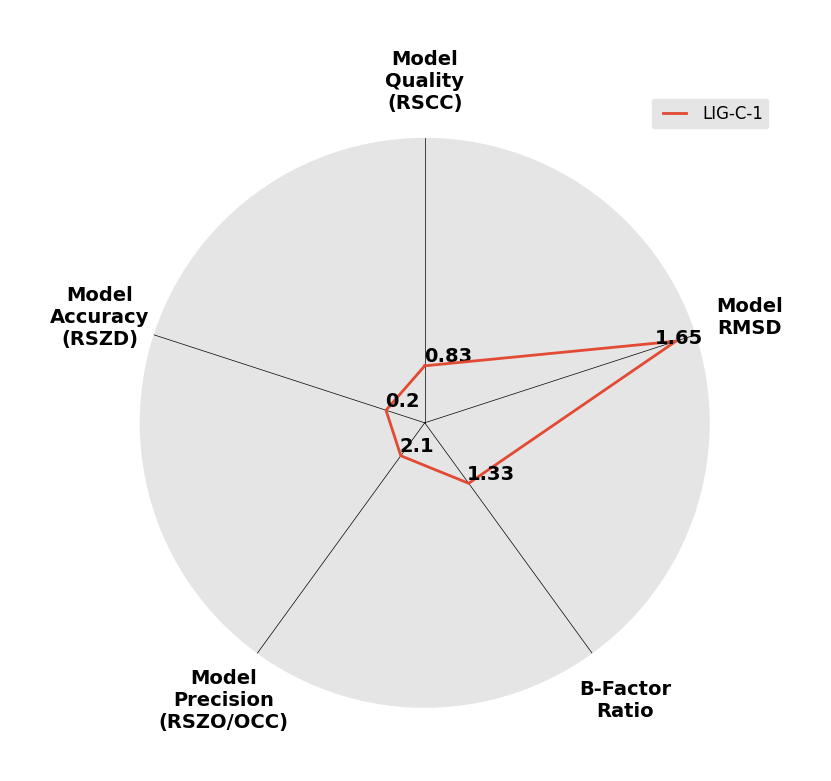
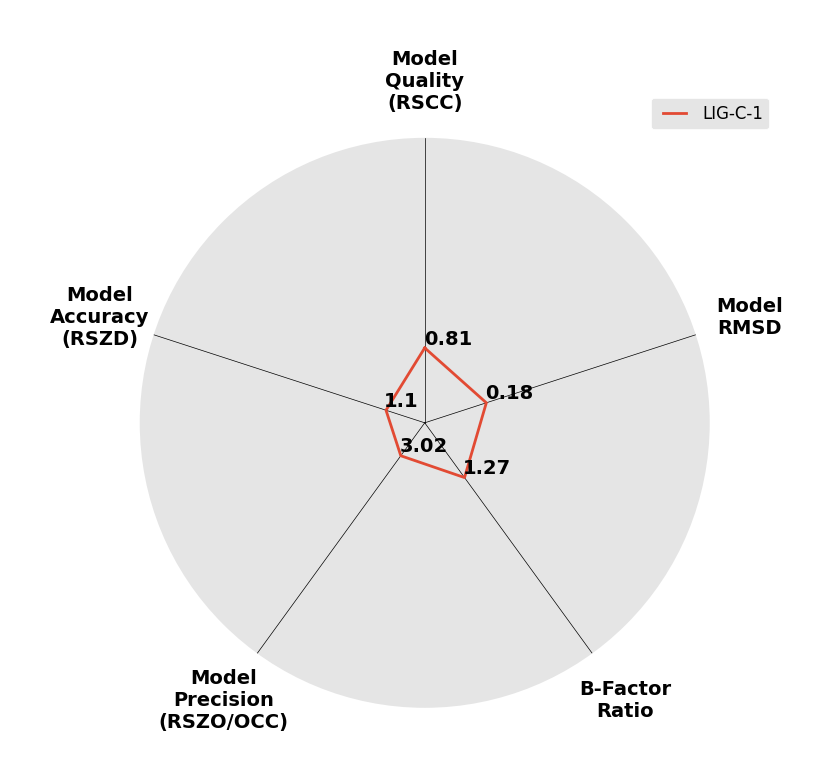
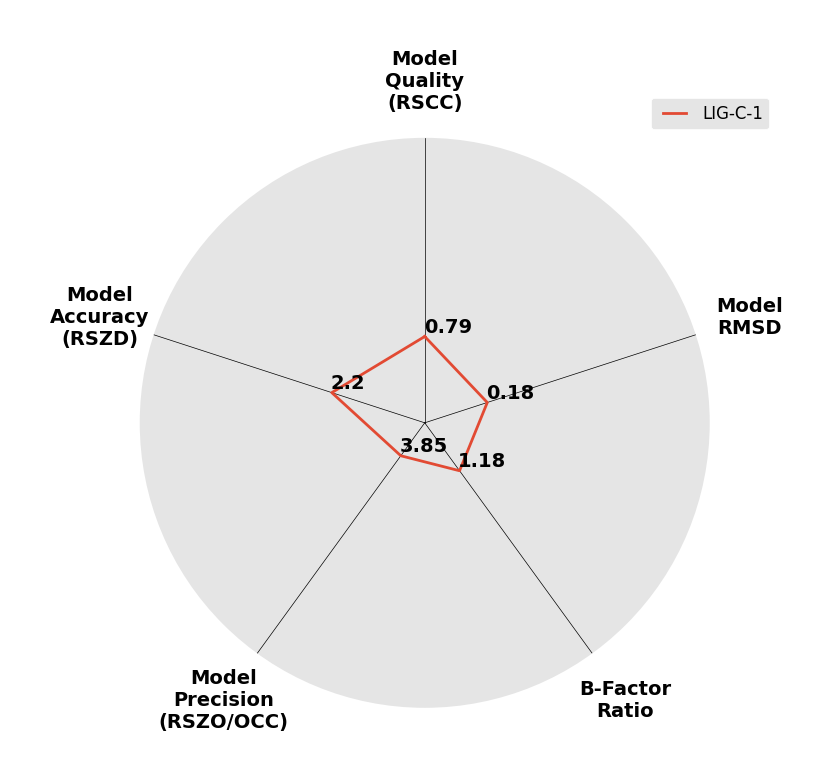
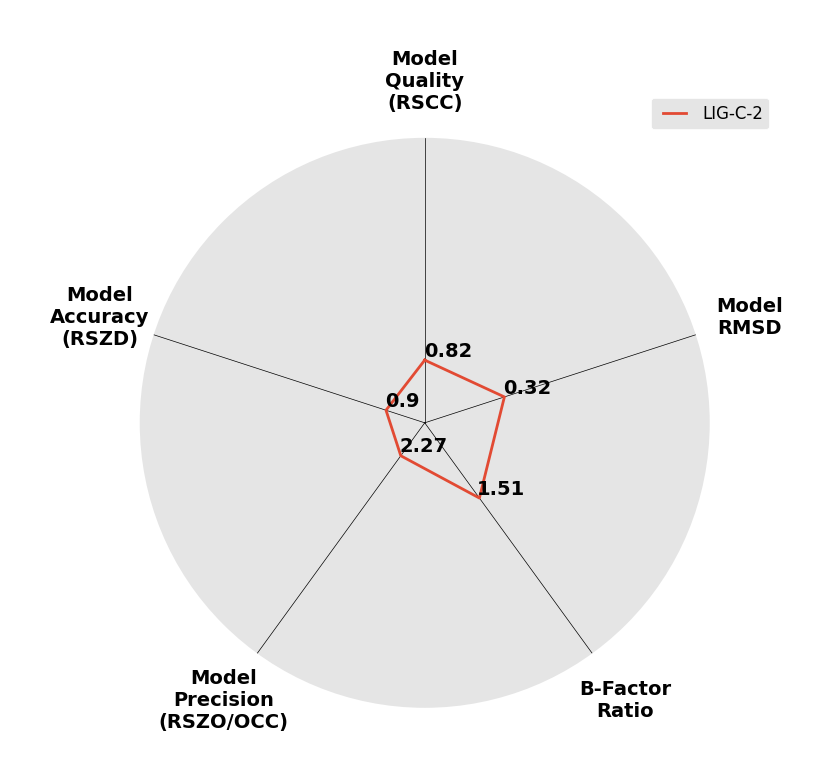
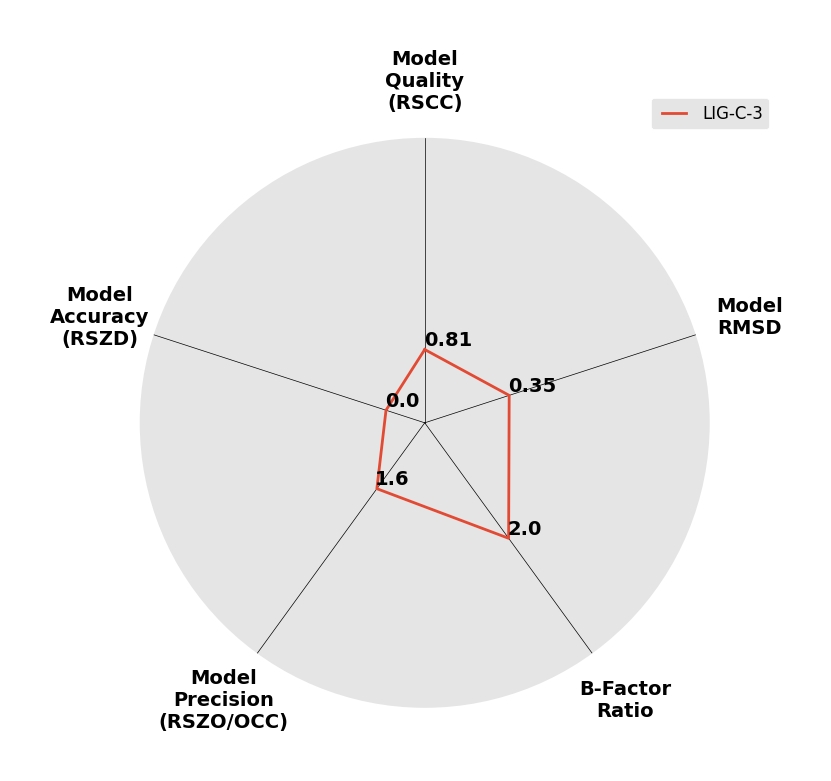
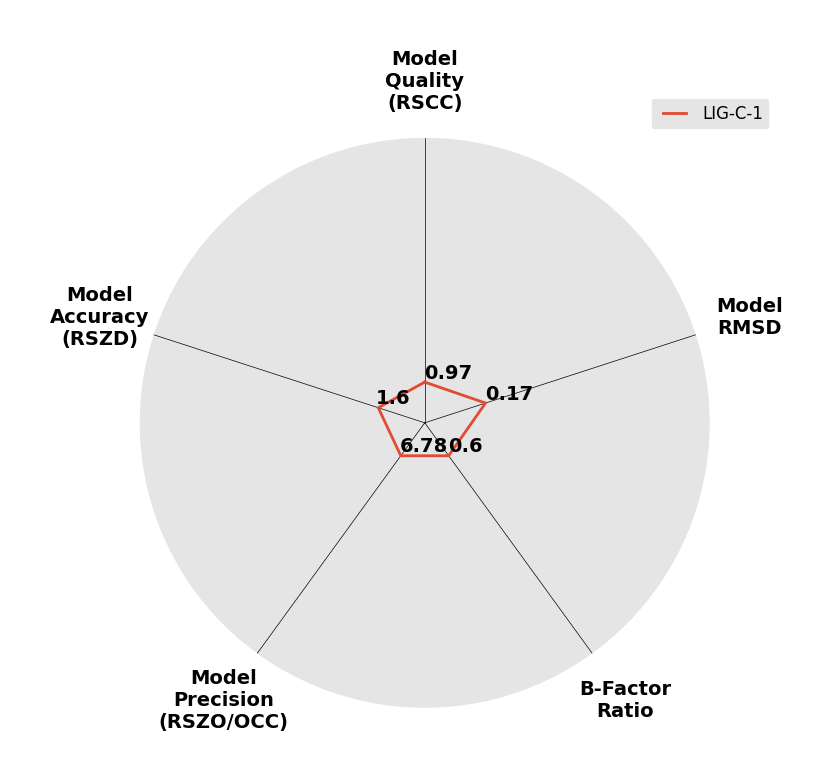
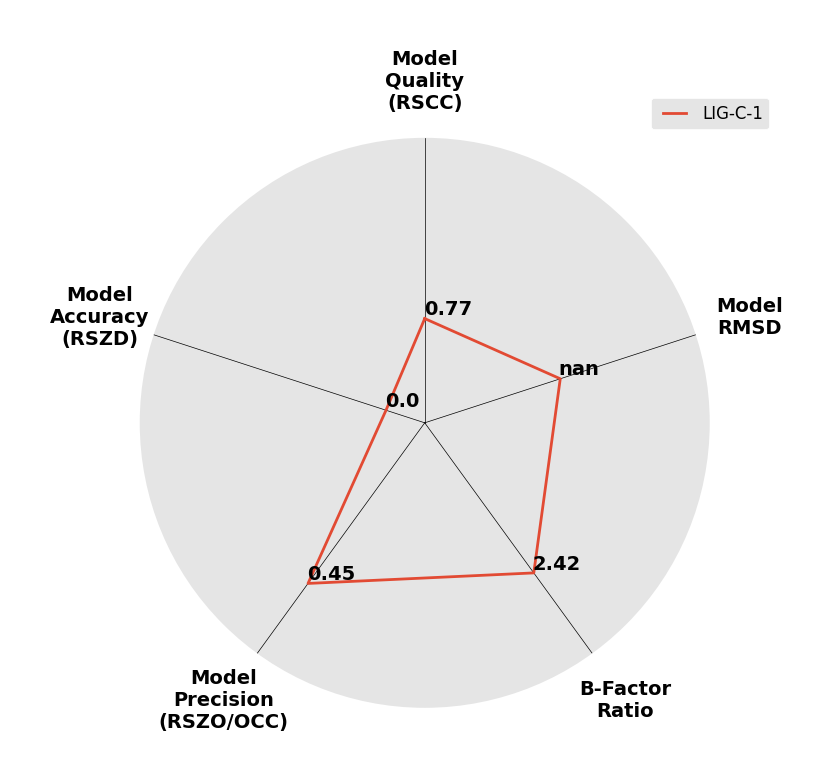
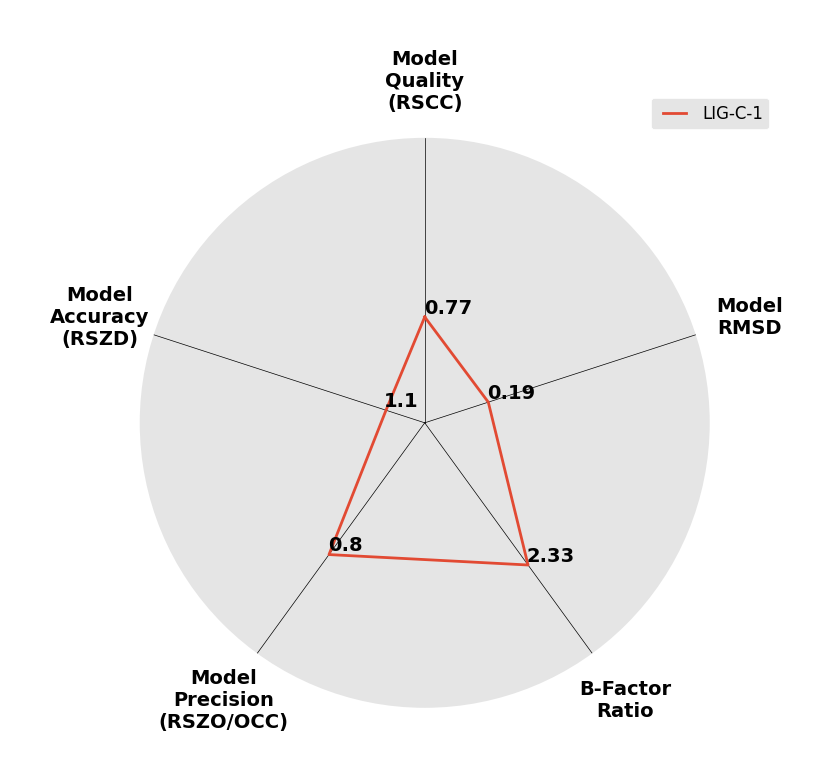
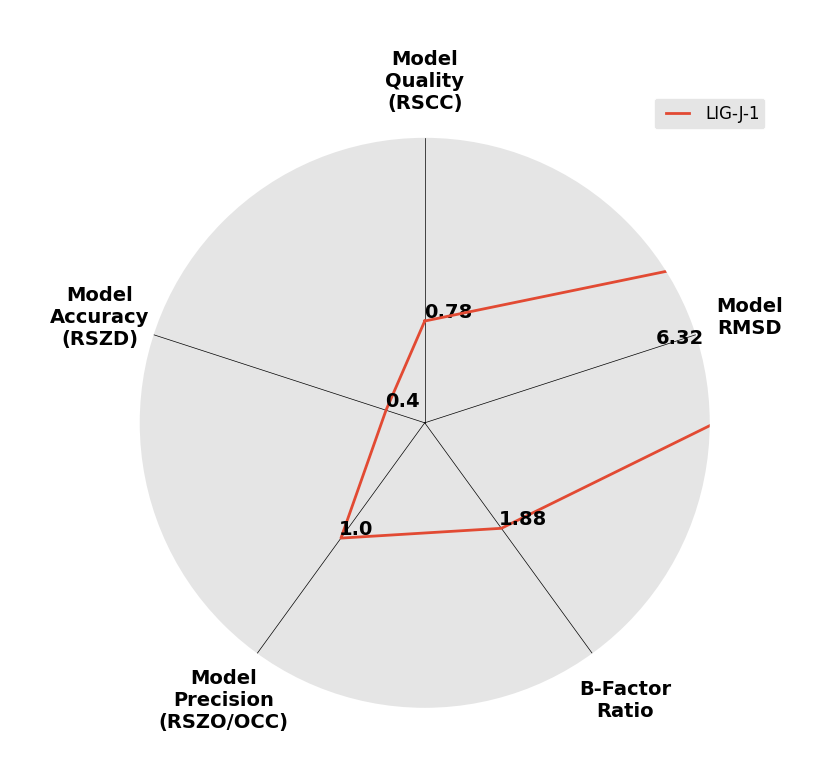
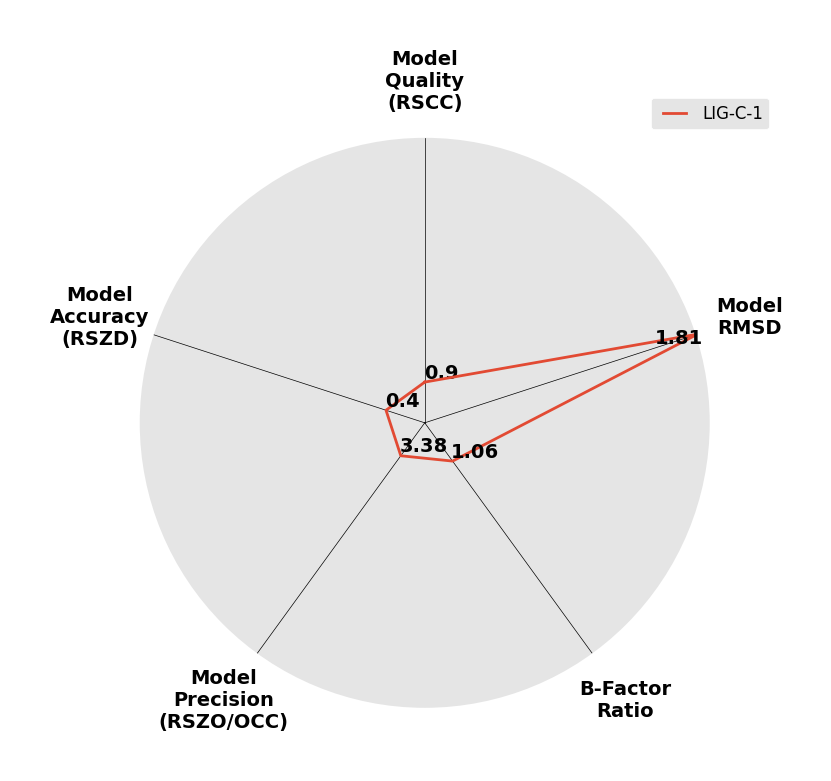
Interpreting 'Ligand validation' spider plots:
Each axis represents one of the values described below; small is better, and large values on any axis implies that further investigation is warranted.Quality (RSCC) reflects the fit of the atoms to the experimental density, and should typically be greater than 0.7.
Accuracy (RSZD) measures the amount of difference density that is found around these atoms, and should be below 3.
B-factor ratio measures the consistency of the model with surrounding protein, and is calculated from the B factors of respectively the changed atoms and all side-chain atoms within 4Å. Large values (>3) reflect poor evidence for the model, and intermediate values (1.5+) indicate errors in refinement or modelling; for weakly-binding ligands, systematically large ratios may be justifiable.
RMSD compares the positions of all atoms built into event density, with their positions after final refinement, and should be below 1Å.
Precision (RSZO/OCC) measures how clear the density is after refinement. (This is not a quality indicator, but is related to strength of binding but not in a straightforward way.)
| Crystal ID | PDB ID | Ligand ID | Ligand confidence | Model status | Compound | Ligand Validation | Event Map 3D | Resol | SPG/ Cell | Files |
|---|---|---|---|---|---|---|---|---|---|---|
| DCLRE1AA-x0128 | 5Q1J | LIG-C-1 | Low | 6 - Deposited |  |
 |
1.42 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x0135 | 5Q1K | LIG-C-1 | 4 - High confidence | 6 - Deposited | 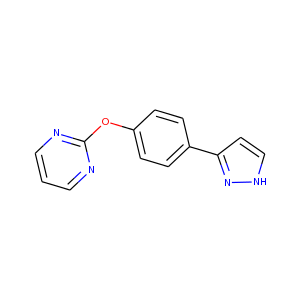 |
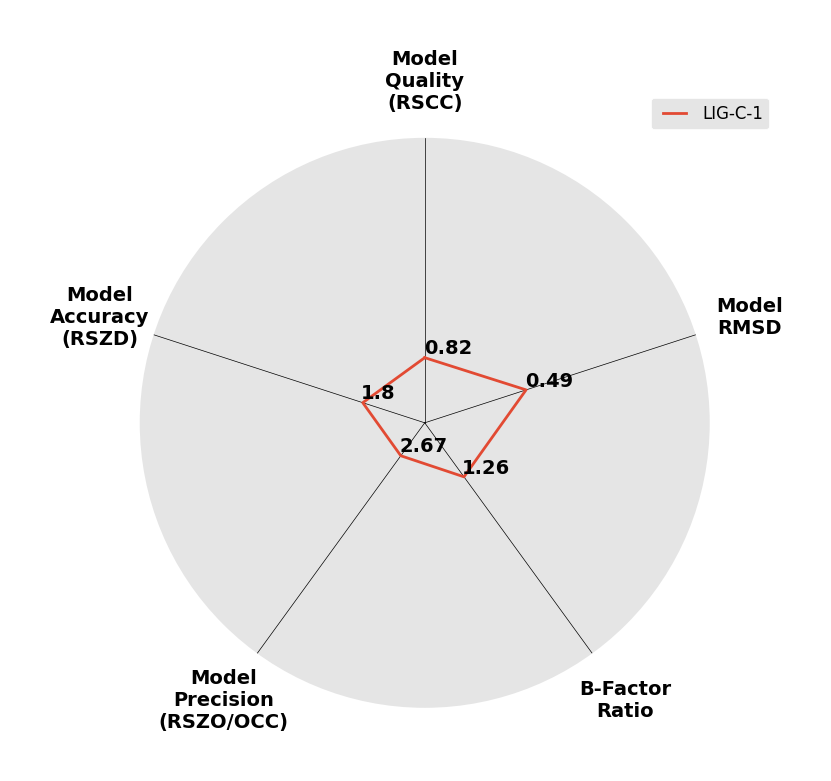 |
1.42 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x0139 | 5Q1L | LIG-C-1 | Low | 6 - Deposited | 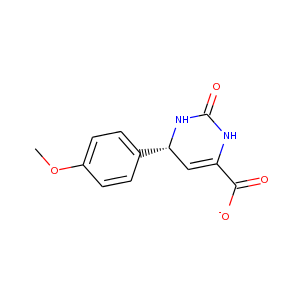 |
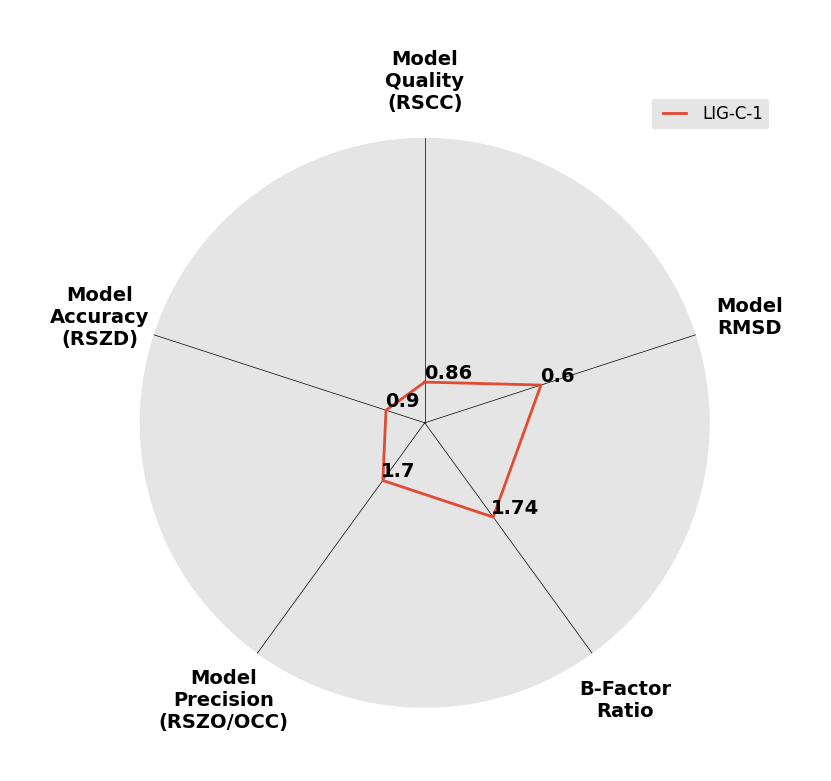 |
1.76 | P 21 21 2152 57 114 90 90 90 | Save | |
| DCLRE1AA-x0150 | 5Q1M | LIG-C-1 | Medium | 6 - Deposited | 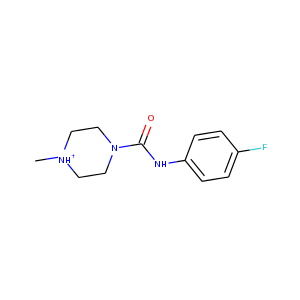 |
 |
1.56 | P 21 21 2151 56 115 90 90 90 | Save | |
| DCLRE1AA-x0157 | 5Q1N | LIG-C-1 | Low | 6 - Deposited | 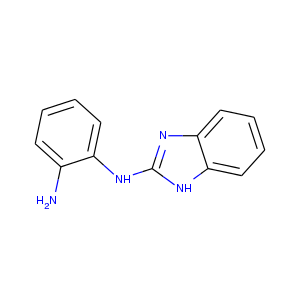 |
 |
1.38 | P 21 21 2152 56 115 90 90 90 | Save | |
| DCLRE1AA-x0164 | 5Q1O | LIG-C-1 | Low | 6 - Deposited |  |
 |
1.25 | P 21 21 2151 57 115 90 90 90 | Save | |
| DCLRE1AA-x0169 | 5Q1P | LIG-C-1 | Low | 6 - Deposited |  |
 |
1.56 | P 21 21 2151 57 114 90 90 90 | Save | |
| DCLRE1AA-x0172 | 5Q1Q | LIG-C-1 | 4 - High Confidence | 6 - Deposited | 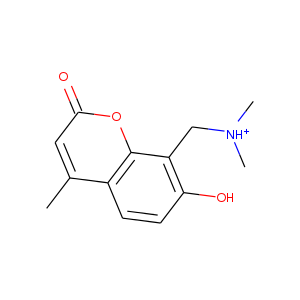 |
 |
1.44 | P 21 21 2152 56 114 90 90 90 | Save | |
| DCLRE1AA-x0183 | 5Q1R | LIG-C-1 | Low | 6 - Deposited | 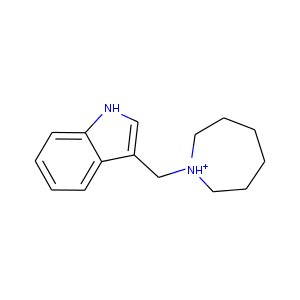 |
 |
1.49 | P 21 21 2151 56 115 90 90 90 | Save | |
| DCLRE1AA-x0193 | 5Q1S | LIG-C-1 | Low | 6 - Deposited |  |
 |
1.62 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x0194 | 5Q1T | LIG-C-1 | Low | 6 - Deposited |  |
 |
1.49 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x0195 | 5Q1U | LIG-C-1 | Medium | 6 - Deposited | 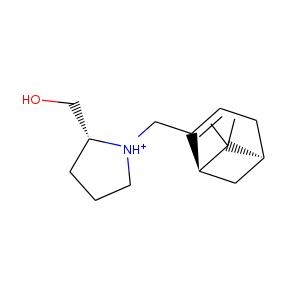 |
 |
1.50 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x0209 | 5Q1V | LIG-C-1 | 2 - Correct Ligand, Weak Density | 6 - Deposited | 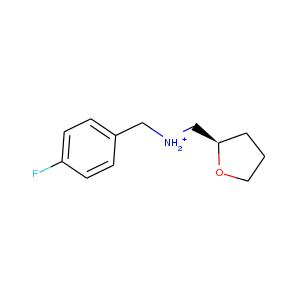 |
 |
1.57 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x0233 | 5Q1W | LIG-C-1 | 4 - High Confidence | 6 - Deposited |  |
 |
1.68 | P 21 21 2152 57 114 90 90 90 | Save | |
| DCLRE1AA-x0233 | 5Q1W | LIG-C-2 | Medium | 6 - Deposited |  |
 |
1.68 | P 21 21 2152 57 114 90 90 90 | Save | |
| DCLRE1AA-x0233 | 5Q1W | LIG-C-3 | Medium | 6 - Deposited |  |
 |
1.68 | P 21 21 2152 57 114 90 90 90 | Save | |
| DCLRE1AA-x0234 | 5Q1X | LIG-C-1 | Low | 6 - Deposited |  |
 |
1.42 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x0234 | 5Q1X | LIG-C-2 | Low | 6 - Deposited |  |
 |
1.42 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x0234 | 5Q1X | LIG-C-3 | not assigned | 6 - Deposited |  |
 |
1.42 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x0928 | 5Q1Z | LIG-C-1 | 4 - High Confidence | 6 - Deposited |  |
 |
1.56 | P 21 21 2152 56 115 90 90 90 | Save | |
| DCLRE1AA-x0965 | 5Q20 | LIG-C-1 | 4 - High Confidence | 6 - Deposited |  |
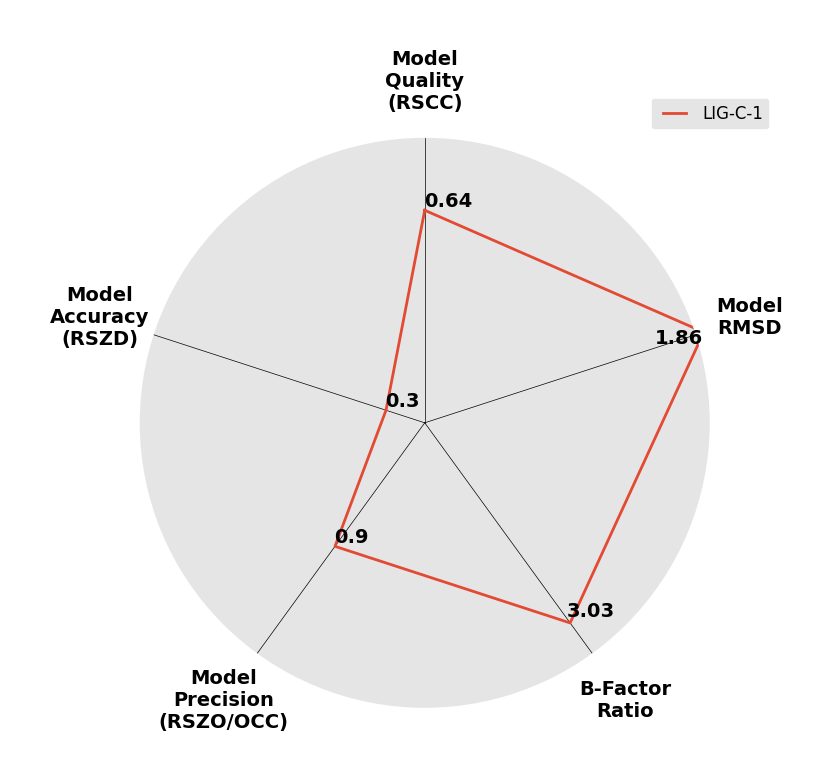 |
1.95 | P 21 21 2151 56 114 90 90 90 | Save | |
| DCLRE1AA-x0972 | 5Q23 | LIG-C-1 | 4 - High Confidence | 6 - Deposited |  |
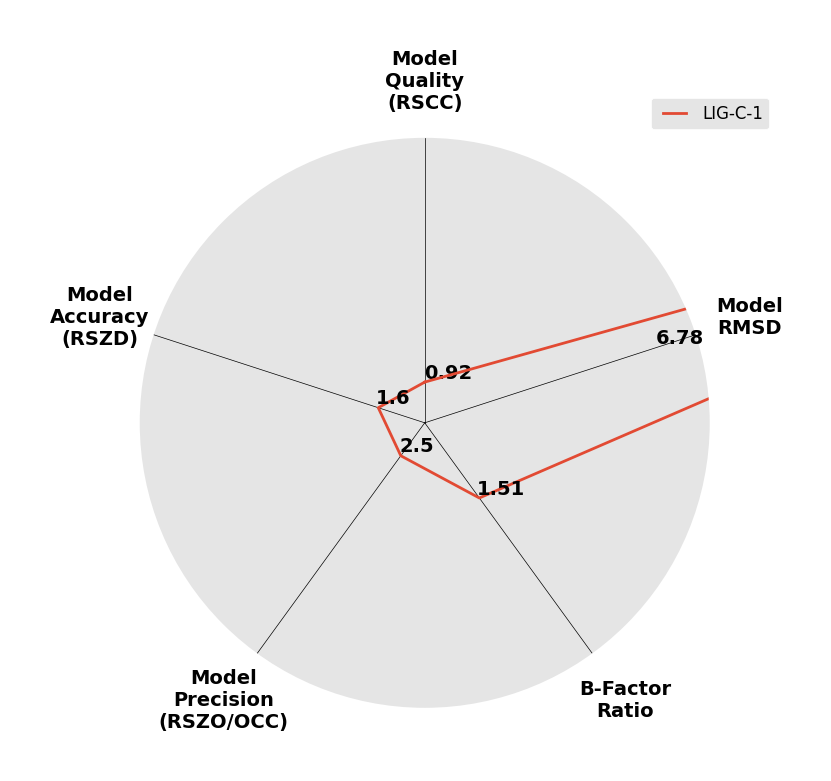 |
1.47 | P 21 21 2152 57 115 90 90 90 | Save | |
| DCLRE1AA-x1010 | 5Q24 | LIG-C-1 | 4 - High Confidence | 6 - Deposited |  |
 |
1.51 | P 21 21 2151 57 114 90 90 90 | Save | |
| DCLRE1AA-x1012 | 5Q25 | LIG-C-1 | 4 - High Confidence | 6 - Deposited |  |
 |
1.60 | P 21 21 2151 56 114 90 90 90 | Save | |
| DCLRE1AA-x1013 | 5Q26 | LIG-C-1 | 4 - High Confidence | 6 - Deposited |  |
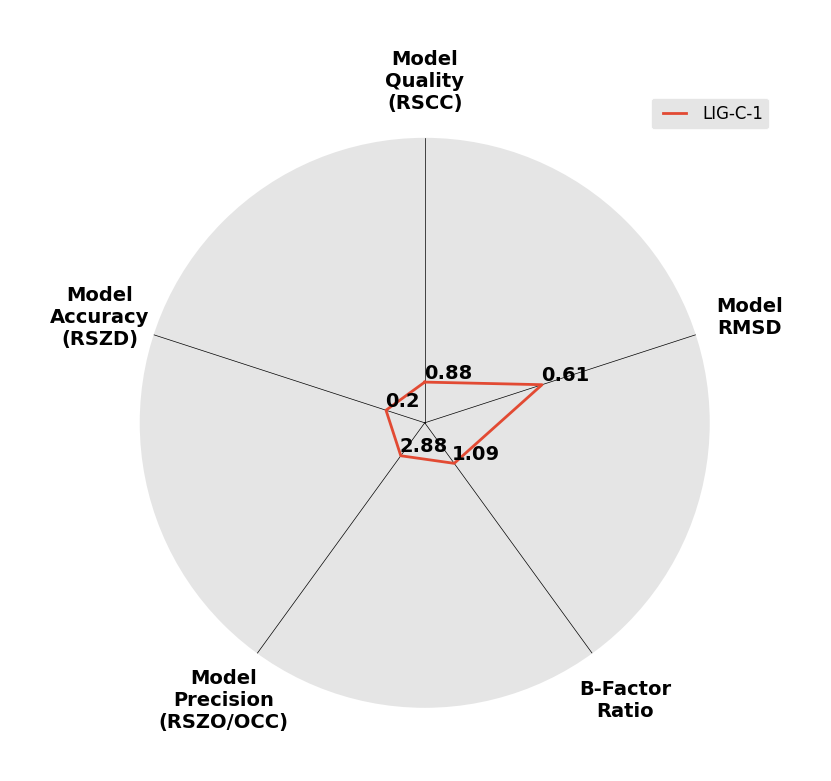 |
1.85 | P 21 21 2151 56 115 90 90 90 | Save | |
| DCLRE1AA-x1015 | 5Q27 | LIG-J-1 | 4 - High Confidence | 6 - Deposited |  |
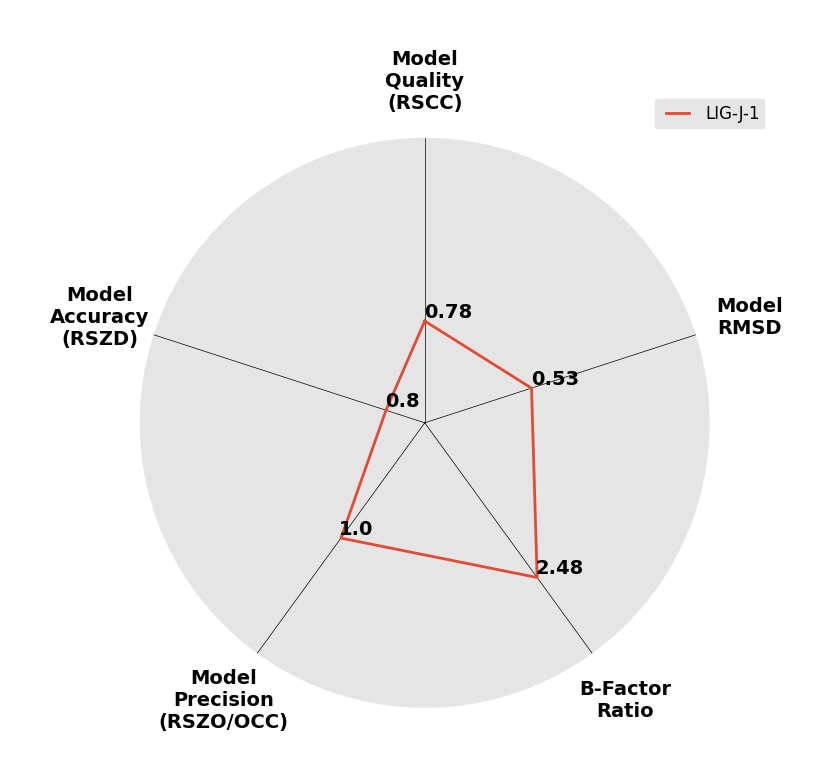 |
1.83 | P 21 21 2151 57 115 90 90 90 | Save | |
| DCLRE1AA-x1074 | None | LIG-J-1 | Low | 4 - CompChem ready |  |
 |
1.66 | P 21 21 2151 56 114 90 90 90 | Save | |
| DCLRE1AA-x1085 | 5Q28 | LIG-C-1 | Medium | 6 - Deposited |  |
 |
1.73 | P 21 21 2151 56 114 90 90 90 | Save |