
Dr. Dalia Barsyte-Lovejoy, PhD is an Assistant Professor at the Department of Pharmacology and Toxicology, UofT, and Principal Investigator at the SGC-Toronto, working to understand fundamental regulatory mechanisms of epigenetic proteins and their pharmacological modulation in cancer. The group’s research focuses on disease mechanisms, therapeutic targets, and chemical probe discovery, resulting in over 30 extensively characterized compounds that have helped shape the emerging field of epigenetics and enabled over 50 collaborative projects that are uncovering new epigenetic mechanisms in cancer and its treatment.
We are interested in understanding the mechanism of epigenetic regulators and posttranslational modifications that control cancer cell growth, differentiation, and therapy response. Protein lysine and arginine methyltransferases regulate transcription, genome stability, splicing, RNA metabolism, and other cell processes dictated by which substrates these enzymes methylate. Lysine methyltransferases such as EZH2 and NSD2 primarily methylate histones to establish repressive and active chromatin. In contrast, arginine methyltransferases have a broad scope of substrates ranging from histones to signaling molecules, enzymes, and structural proteins. Epigenetic chromatin regulation, transcriptome, and cellular signaling are fine-tuned by ubiquitin modification. Our work seeks to understand how these posttranslational modifications are misregulated in cancer and identify new therapeutic targets.
Through multidisciplinary research that includes cell and chemical biology, protein structural biology, and many collaborative studies with colleagues across industry and academia, the SGC chemical probes project has generated several probes for methyltransferases, ubiquitin ligases, and deubiquitylases. We are currently using these chemical probes to explore the cellular pathways in poor prognosis acute myeloid leukemia, pancreatic, lung and breast cancer.
Chemical probes as tools for cancer target discovery
|
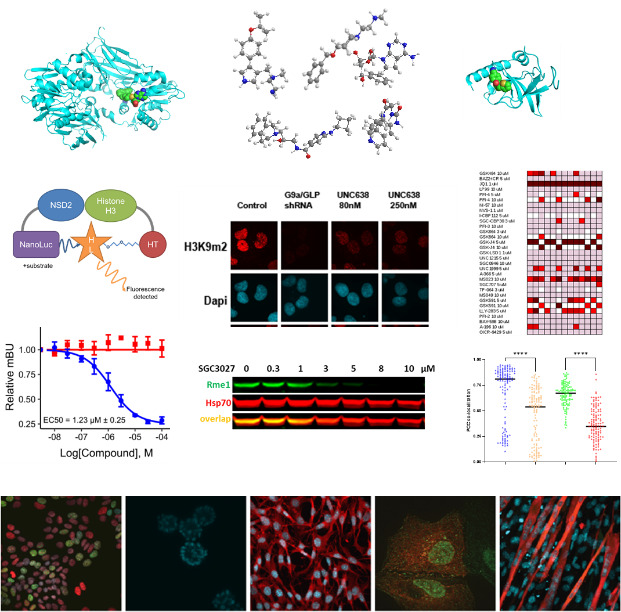
To study epigenetic modifier proteins, we need genetic and pharmacological tools. Chemical probe compounds that potently and selectively inhibit or degrade the target proteins in cells provide tools for modulating activating/repressing histone marks and other cellular signaling pathways. By discovering and using chemical probes, we expand our understanding of the protein function and its therapeutic utility to establish a biological rationale in cancer therapy.
|
Link to Open Lab notebooks that features science community posts on our various projects https://openlabnotebooks.org/
Righetto GL, Yin Y, Duda DM, Vu V, Szewczyk MM, Zeng H, Li Y, Loppnau P, Mei T, Li YY, Seitova A, Patrick AN, Brazeau JF, Chaudhry C, Barsyte-Lovejoy D, Santhakumar V, Halabelian L
PNAS Nexus. 2024-4-26 . 3(4):pgae153 .doi: 10.1093/pnasnexus/pgae153
PMID: 38665159Shen L, Ma X, Wang Y, Wang Z, Zhang Y, Pham HQH, Tao X, Cui Y, Wei J, Lin D, Abeywanada T, Hardikar S, Halabelian L, Smith N, Chen T, Barsyte-Lovejoy D, Qiu S, Xing Y, Yang Y
Nat Commun. 2024-4-1 . 15(1):2809 .doi: 10.1038/s41467-024-47107-9
PMID: 38561334Yazdi AK, Perveen S, Dong C, Song X, Dong A, Szewczyk MM, Calabrese MF, Casimiro-Garcia A, Chakrapani S, Dowling MS, Ficici E, Lee J, Montgomery JI, O'Connell TN, Skrzypek GJ, Tran TP, Troutman MD, Wang F, Young JA, Min J, Barsyte-Lovejoy D, Brown PJ, Santhakumar V, Arrowsmith CH, Vedadi M, Owen DR
RSC Med Chem. 2024-3-20 . 15(3):1066-1071 .doi: 10.1039/d3md00633f
PMID: 38516600Kimani SW, Perveen S, Szewezyk M, Zeng H, Dong A, Li F, Ghiabi P, Li Y, Chau I, Arrowsmith CH, Barsyte-Lovejoy D, Santhakumar V, Vedadi M, Halabelian L
Commun Biol. 2023-12-16 . 6(1):1272 .doi: 10.1038/s42003-023-05655-8
PMID: 38104184Fnaiche A, Chan HC, Paquin A, González Suárez N, Vu V, Li F, Allali-Hassani A, Cao MA, Szewczyk MM, Bolotokova A, Allemand F, Gelin M, Barsyte-Lovejoy D, Santhakumar V, Vedadi M, Guichou JF, Annabi B, Gagnon A
ACS Med Chem Lett. 2023-12-14 . 14(12):1746-1753 .doi: 10.1021/acsmedchemlett.3c00386
PMID: 38116405Alqazzaz MA, Luciani GM, Vu V, Machado RAC, Szewczyk MM, Adamson EC, Cheon S, Li F, Arrowsmith CH, Minden MD, Barsyte-Lovejoy D
Exp Hematol. 2023-12-10 . 104135 .doi: 10.1016/j.exphem.2023.11.009
PMID: 38072134Fnaiche A, Mélin L, Suárez NG, Paquin A, Vu V, Li F, Allali-Hassani A, Bolotokova A, Allemand F, Gelin M, Cotelle P, Woo S, LaPlante SR, Barsyte-Lovejoy D, Santhakumar V, Vedadi M, Guichou JF, Annabi B, Gagnon A
Bioorg Med Chem Lett. 2023-9-26 . 129488 .doi: 10.1016/j.bmcl.2023.129488
PMID: 37770003Ramachandran S, Szewczyk M, Barghout SH, Ciulli A, Barsyte-Lovejoy D, Vu V
Methods Mol Biol. 2023-8-10 . 2706:149-165 .doi: 10.1007/978-1-0716-3397-7_11
PMID: 37558947Szewczyk MM, Owens DDG, Barsyte-Lovejoy D
Methods Mol Biol. 2023-8-10 . 2706:137-148 .doi: 10.1007/978-1-0716-3397-7_10
PMID: 37558946Harding RJ, Franzoni I, Mann MK, Szewczyk MM, Mirabi B, Ferreira de Freitas R, Owens DDG, Ackloo S, Scheremetjew A, Juarez-Ornelas KA, Sanichar R, Baker RJ, Dank C, Brown PJ, Barsyte-Lovejoy D, Santhakumar V, Schapira M, Lautens M, Arrowsmith CH
J Med Chem. 2023-7-27 . .doi: 10.1021/acs.jmedchem.3c00314
PMID: 37499118