 |
 |
 |
 |
 |
 |

Protein tyrosine phosphatases (PTPs) play a critical role regulating cell signaling by selectively dephosphorylating their substrates. So far our laboratory has released 22 human high resolution crystal structures which, together with prior structural knowledge, results in an excellent structural coverage of the classical PTP family. A recent structural analysis carried out in our laboratory compared the existing structural information available for this enzyme family [1]. The comparison revealed that despite their largely conserved fold, surface properties of PTPs are strikingly diverse. A number of unique and shared features has been identified that offers an excellent basis for structure-based design efforts of target specific inhibitors. Many PTPs have been recognized as potential targets [2] for the development of new therapies and the development of selective inhibitors is an ongoing effort in our laboratory.
1. Barr et al.. Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome. Cell (2009) 136(2):352-363.
2. Tautz et al.. Targeting the PTPome in human disease. Expert Opin. Ther. Targets. (2006) 10(1):157-77.
3. Tonks. Protein Tyrosine Phosphatases: From Gene to Function to Disease. Nat Rev Mol Cell Biol. (2006) 7(11):833-846.
About the image: Similar to early comparisons of different species that led to the establishment of phylogenetic trees, high resolution structural information can be used to identify unique as well as shared structural properties that can be explored and cluster members of a enzyme family into a tree based on molecular properties. The structural comparison of PTPs revealed that many features cannot be reliably predicted from sequence analysis alone. The phosphatase images are coloured according to the surface electrostatic potential using ICM.
PTP Domain Structural Coverage
The human genome contains 107 PTPs grouped into four distinct families: Class I cysteine-based PTPs constitute the largest family. They are divided into 38 “classical” tyrosine specific PTPs and 61 dual specificity phosphatases (DUSPs) [4]. The classical PTPs are one of the most comprehensively structurally covered protein families: 32 structures of the 49 D1/D2 domains are currently available. PTPs share a highly conserved overall fold but have very diverse protein surface properties.
4. Alonso et al.. Protein tyrosine phosphatases in the human genome. Cell (2004) 117(6):699-711.
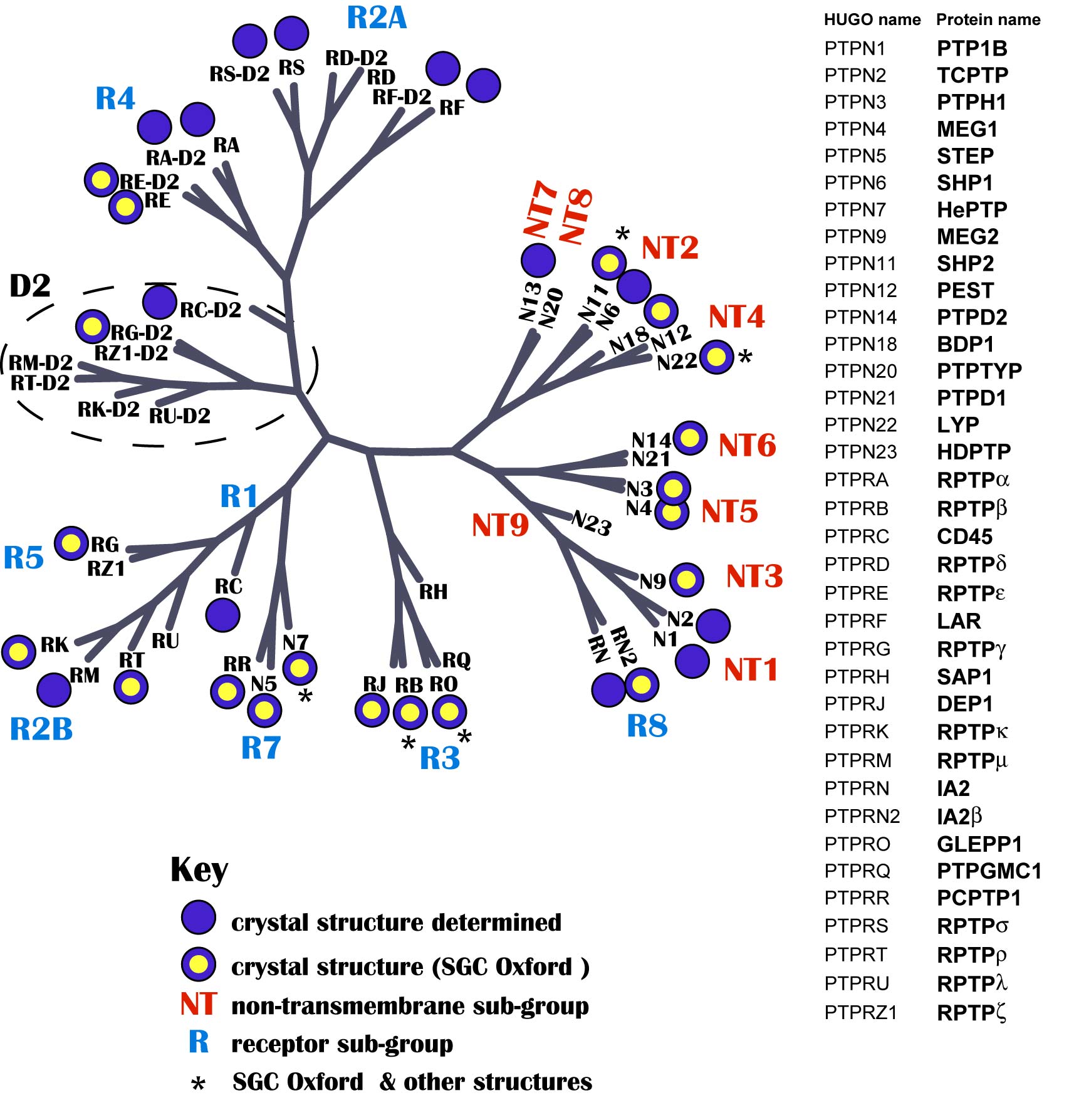
Structural coverage of the human PTP family: Catalytic domains with known three dimensional structure are highlighted by a dot. Dots with a yellow center indicate structures released by our laboratory.
Recent human phosphatase domain structure depositions in the PDB
PDB code | Release date | Phosphatase | Species | Description | Sector |
|---|---|---|---|---|---|
| 3EU0 | 11.Nov.2008 | PTPN1 | Human | Crystal structure of the S-nitrosylated Cys215 of PTP1B | Academia |
| 2ZMM | 07.Oct.2008 | PTPN1 | Human | Crystal structure of PTP1B-inhibitor complex, 4-bromo-3-(carboxymethoxy)- 5-{3-[cyclohexyl (methylcarbamoyl) amino]phenyl} thiophene-2- carboxylic acid | Industry |
| 2ZN7 | 07.Oct.2008 | PTPN1 | Human | Crystal structure of PTP1B-inhibitor complex, 4-bromo-3-(carboxymethoxy)- 5-{3-[cyclohexyl (phenylcarbonyl) amino]phenyl} thiophene-2- carboxylic acid | Industry |
| 3D9C | 23.Sep.2008 | PTPN1 | Human | Crystal Structure PTP1B complex with aryl Seleninic acid | Academia |
| 2QDC | 24.Jun.2008 | PTPN7 | Human | Crystal structure of the HePTP catalytic domain D236A mutant | Academia |
| 2QDM | 24.Jun.2008 | PTPN7 | Human | Crystal structure of the HePTP catalytic domain C270S/D236A/Q314A mutant | Academia |
| 2QDP | 24.Jun.2008 | PTPN7 | Human | Crystal structure of the HePTP catalytic domain C270S mutant crystallized in ammonium acetate | Academia |
| 3CWE | 10.Jun.2008 | PTPN1 | Human | PTP1B in complex with a phosphonic acid inhibitor | Industry |
| 2JJD | 08.Apr.2008 | PTPRE | Human | pProtein tyrosine phosphatase, receptor type, E isoform | SGC Oxford |
PTP Structures deposited by SGC
Click on the protein name to read more about the phosphatase and its structure in our website. To explore and view the structure in your web browser, please click on the "iSee" icon (first-time user: you'll be asked to install our visualisation plug-in - please follow the installation instructions that will be displayed).
HUGO name | Protein Name | PDB code | ||
 | RPTPκ | 2C7S |  | |
 | RPTPρ | 2OOQ |  | |
 | RPTPε | 2JJD | coming soon | |
 | RPTPγ | 2NLK |  | |
 | RPTPγ | 2H4V |  | |
 | RPTPβ | 2AHS |  | |
 | DEP1 | 2CFV, 2NZ6 |  | |
 | GLEPP1 | 2GJT |  | |
 | PCPTP1 | 2A8B |
| |
 | STEP | 2BIJ |  | |
 | STEP | 2CJZ |  | |
 | STEP | 2BV5 |  |
HUGO name | Protein Name | PDB code | ||
 | HePTP | 2A3K |  | |
 | IA2β | 2QEP |  | |
 | SHP2 | 3B7O |  | |
 | MEG2 | 2PA5 |  | |
 | PTPH1 | 2B49 |  | |
 | MEG1 | 2I75 |  | |
 | BDP1 | 2OC3 |  | |
 | LYP | 2P6X |  | |
 | PTPD2 | 2BZL |  |
PTP-related SGC publications
Original articles
- Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome. Alastair J. Barr, Emilie Ugochukwu, Wen Hwa Lee, Oliver King, Panagiotis Filippakopoulos, Ivan Alfano, Pavel Savitsky, Nicola Burgess-Brown, Susanne Müller and Stefan Knapp. Cell (2009).
- MAP-kinase specific tyrosine phosphatases (PTPN5, PTPN7 and PTPRR) – New targets for drug discovery? Alastair J. Barr and Stefan Knapp. Trends Pharmacol Sci. (2006) 27(10):525-530.
- The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1. Jeyanthy Eswaran, Judit É. Debreczeni, Emma Longman, Alastair J. Barr and Stefan Knapp. Protein Science. (2006) 15(6):1500-1505.
- Crystal Structure of Human Protein Tyrosine Phosphatase 14 (PTPN14) at 1.65 Å resolution. Alastair J. Barr, Judit É. Debreczeni, Jeyanthy Eswaran and Stefan Knapp. Proteins. (2006) 63 (4):1132-1136.
- Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7 - a family of human MAP-kinase specific protein tyrosine phosphatases. Jeyanthy Eswaran, Jens Peter von Kries, Brian Marsden, Emma Longman, Judit É. Debreczeni, Emilie Ugochukwu, Andrew Turnbull, Wen Hwa Lee, Stefan Knapp and Alastair J Barr. Biochem J. (2006) 395(3):483-491.
