| Probe | Negative control | |
 |
|  |
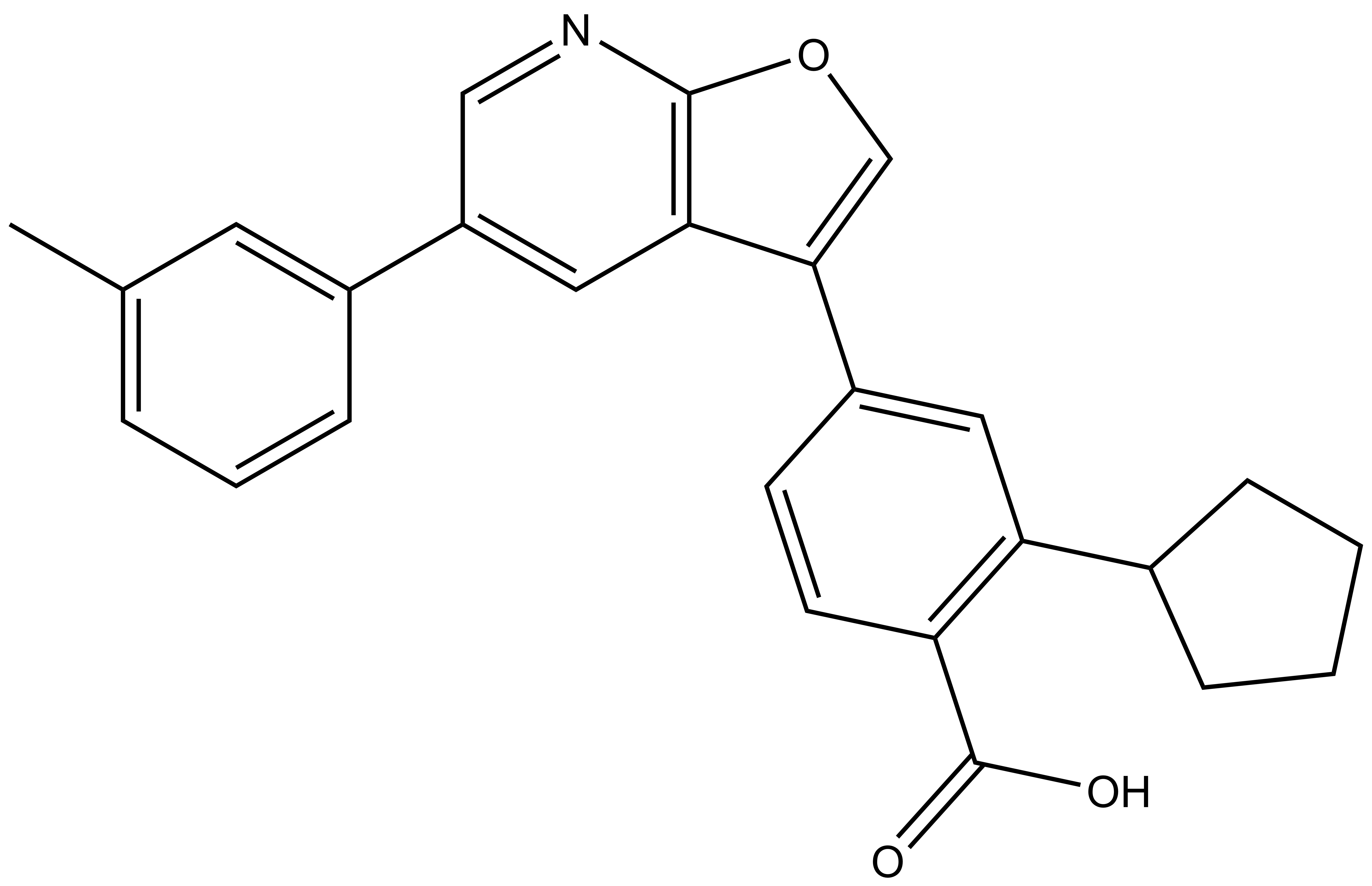
SGC-CAMKK2-1 |
| SGC-CAMKK2-1N |
Chemical structures of furopyridines SGC-CAMKK2-1 (CAMKK2/CAMKK1 chemical probe) and SGC-CAMKK2-1N (CAMKK2/CAMKK1 negative control)
In collaboration with scientists at St. Vincent’s Institute at the University of Melbourne, MD Anderson, Duke University, and Baylor College of Medicine, the SGC has developed a chemical probe for the kinase CAMKK2 and a corresponding closely related negative control compound. Because of the active site similarity of CAMKK1 and CAMKK2, it is likely that both targets are engaged. When we have gathered appropriate CAMKK1 data this page will be updated. Until such data becomes available, we recommend that users of the probe consider both CAMKK1 and CAMKK2 as targets.
CAMKK2 is a serine-threonine protein kinase and a member of the “Other” group of protein kinases (Figure 1). CAMKK2 is primarily expressed in the brain, with lower levels in some peripheral tissues including thymus, spleen, lung, and testis.
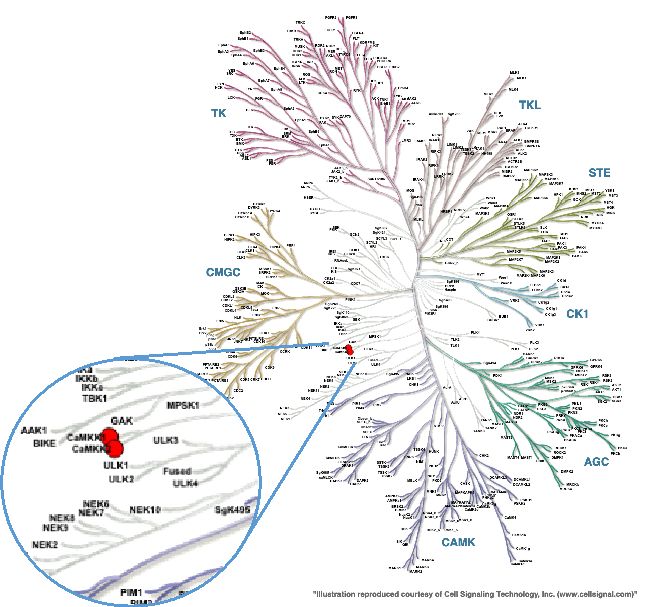
Figure 1. Phylogenetic kinase tree, CAMKK1/CAMKK2 highlighted with red circles. Illustration is reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com).
CAMKK2 is activated by binding to a calcium/calmodulin complex, and activated CAMKK2 can phosphorylate CAMK1, CAMK4, AMPK, and AKT (Figure 2). Activation of these kinases regulates a number of important processes. Because of its involvement in so many critical cellular pathways, CAMKK2 activity has been linked to several diseases. For example, CAMKK2 is upregulated and/or overexpressed in several cancers (Inlcuding gastric, HCC, glioma, and prostate). As such, CAMKK2 inhibitors hold promise as potential therapeutics. STO-609 is used widely as a CAMKK2 inhibitor tool molecule. Although not promiscuous, it has off targets that may confound interpretation of cellular screening results, so a more selective chemical probe or a chemical probe with a different scaffold and different off-target effects will benefit the community.
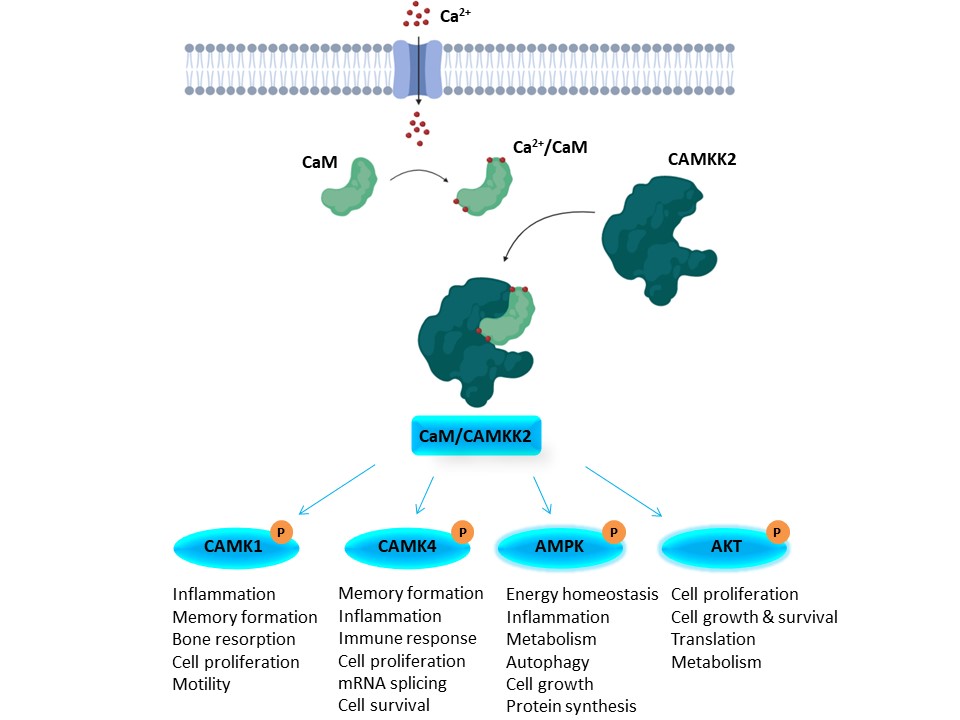
Figure 2. Overview of the CAMKK2 signaling pathway.
SGC-CAMKK2-1N is a suitable structurally similar negative control. It has an IC50 = 27 µM on purified CAMKK2, and cellular potency > 10,000 µM.
| Probe |
 |
SGC-CAMKK2-1 |
| Physical and chemical properties for SGC-CAMKK2-1 | |
| Molecular weight | 397.5 |
| Molecular formula | C26H23NO3 |
| IUPAC name | 2-cyclopentyl-4-(5-(m-tolyl)furo[2,3-b]pyridin-3-yl)benzoic acid |
| MollogP | 5.53 |
| PSA | 63.3 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 4 |
| No. of hydrogen bond acceptors | 4 |
| No. of hydrogen bond donors | 1 |
| Storage | Stable as a solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended. DMSO stock solutions (up to 10mM) are stable at -20 °C |
| Dissolution | Soluble in DMSO up to 10 mM. |
| Negative control |
 |
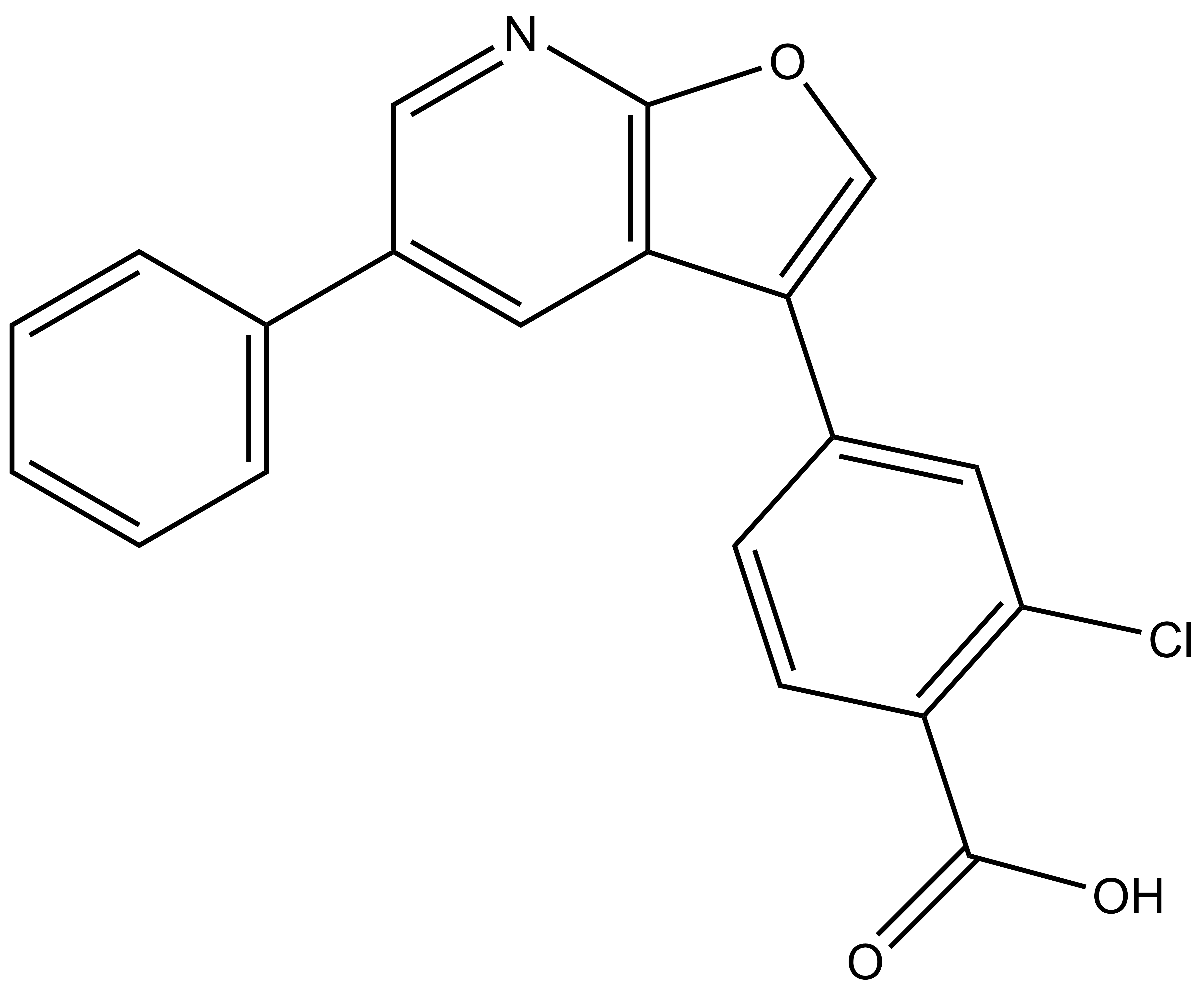
SGC-CAMKK2-1N |
| Physical and chemical properties for SGC-CAMKK2-1N | |
| Molecular weight | 349.8 |
| Molecular formula | C20H12ClNO3 |
| IUPAC name | 2-chloro-4-(5-phenylfuro[2,3-b]pyridin-3-yl)benzoic acid |
| MollogP | 4.37 |
| PSA | 63.33 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 3 |
| No. of hydrogen bond acceptors | 4 |
| No. of hydrogen bond donors | 1 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10mM) are stable at -20 °C. |
| Dissolution | Soluble in DMSO up to 10 mM. |
SMILES:
SGC-CAMKK2-1: CC1=CC=CC(C2=CC3=C(N=C2)OC=C3C4=CC(C5CCCC5)=C(C=C4)C(O)=O)=C1
SGC-CAMKK2-1N: ClC1=C(C=CC(C2=COC3=C2C=C(C=N3)C4=CC=CC=C4)=C1)C(O)=O
InChI:
SGC-CAMKK2-1:1S/C26H23NO3/c1-16-5-4-8-18(11-16)20-13-23-24(15-30-25(23)27-14-20)19-9-10-21(26(28)29)22(12-19)17-6-2-3-7-17/h4-5,8-15,17H,2-3,6-7H2,1H3,(H,28,29)
SGC-CAMKK2-1N: 1S/C20H12ClNO3/c21-18-9-13(6-7-15(18)20(23)24)17-11-25-19-16(17)8-14(10-22-19)12-4-2-1-3-5-12/h1-11H,(H,23,24)
InChIKey:
SGC-CAMKK2-1: TXIYVFVMXNFNRX-UHFFFAOYSA-N
SGC-CAMKK2-1N: KHPDNSBTNDXSBL-UHFFFAOYSA-N
SGC-CAMKK2-1 was profiled in the KINOMEscan assay against 403 wild-type kinases at 1 µM. Only two kinases demonstrated percentage of control (PoC) values of less than 15% (CAMKK2 PoC = 5.4, CAMKK1 PoC = 12), indicating excellent kinome wide selectivity.
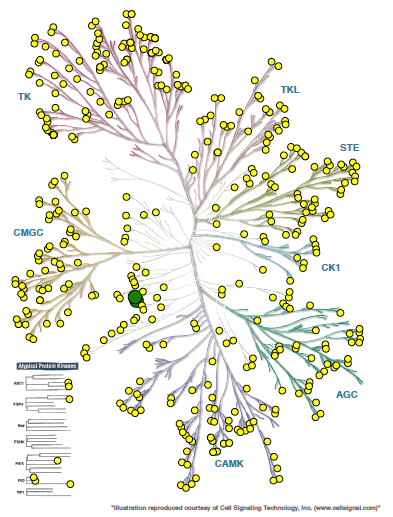
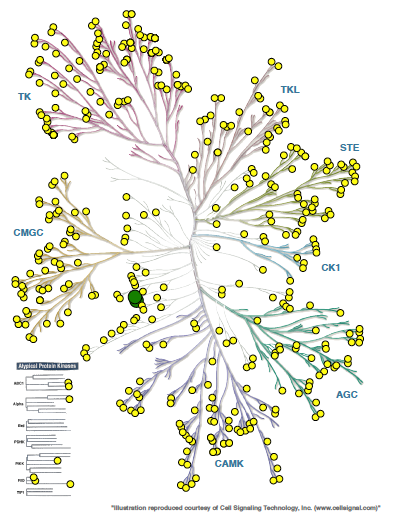
Figure 1: Kinome wide selectivity of SGC-CAMKK2-1. Only CAMKK2 and CAMKK1 bound to the inhibitor with Poc <15 (green dots).
For all other kinases PoC > 15 (yellow dots).
SGC-CAMKK2-1 (also known as YL-36) was tested from 0 – 10 µM in C4-2 cells (a prostate cancer cell line). Expression levels of CaMKK2, p-AMPK and AMPK were measured (Figure 1). The in cell Western IC50 = 1.6 µM.
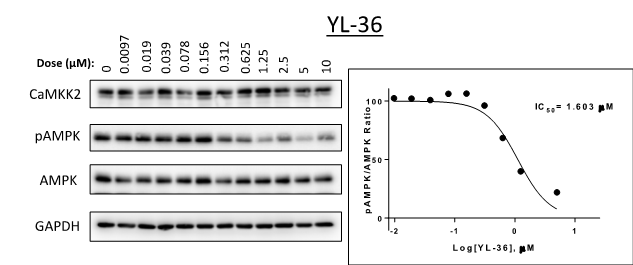
Figure 1: SGC-CAMKK2-1 (also known as YL-36) was tested from 0 – 10 µM in C4-2 cells (a prostate cancer cell line) in western blot assays. Levels of P-AMPK were measured.
This work was performed by members of the Dan Frigo lab at MD Anderson (Dan Frigo, Thomas Pulliam, Chenchu Li, Dominik Awad)
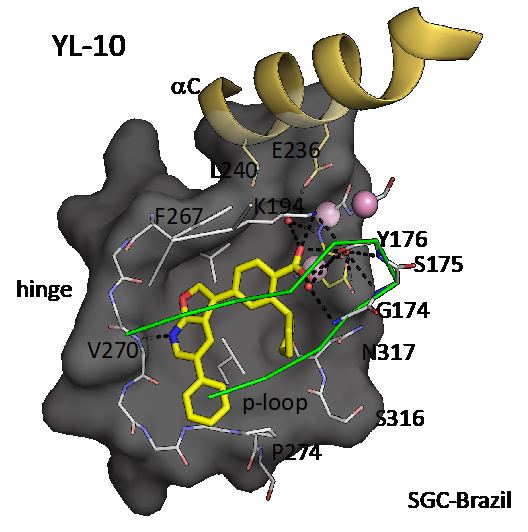
Co-structure of CAMKK2 with YL-10 (resolution 1.7 Å). PDB ID 5UY6. YL-10 is very similar to the probe SGC-CAMKK2-1, only missing a methyl group on the phenyl ring that projects towards solvent.