Target 2035: Accelerating Chemical Probe Discovery Through Open Science
Target 2035 is an open-science global movement comprising scientists and researchers, focusing on the development of molecular tools to study human proteins and inform drug discovery. By fostering collaboration between the public and the private sectors, Target 2035 aims to develop a pharmacological modulator for every protein in the human proteome by 2035.
Why chemical probes matter
Chemical probes are small molecules that help scientists interrogate protein function in living systems. They are essential tools for understanding disease mechanisms, validating new drug targets, and guiding therapeutic development. Yet today, probes exist for only a small portion of the human proteome.
Despite new technologies, the cost and time required to develop these probes have remained largely unchanged over the last decade, particularly for lesser-studied proteins. The current bottleneck is clear: hit-finding - the process of identifying a starting molecule that binds a protein target.
A transformation in how we discover small molecules
Traditional hit-finding is experimental, expensive, and unscalable. Target 2035 aims to shift this process into the computational endeavor by generating vast, high-quality protein–ligand interaction datasets and releasing them openly.
These datasets will enable the training and testing of machine learning models that can predict new binders at scale, thus accelerating the path to probe development.

From stealth mode to global momentum
Between 2020 and 2025, Target 2035 operated in a “stealth mode” phase: building community, mapping challenges, and establishing infrastructure. This foundational work led to the selection of the right technologies, expert teams, and global partnerships to support scaled implementation.
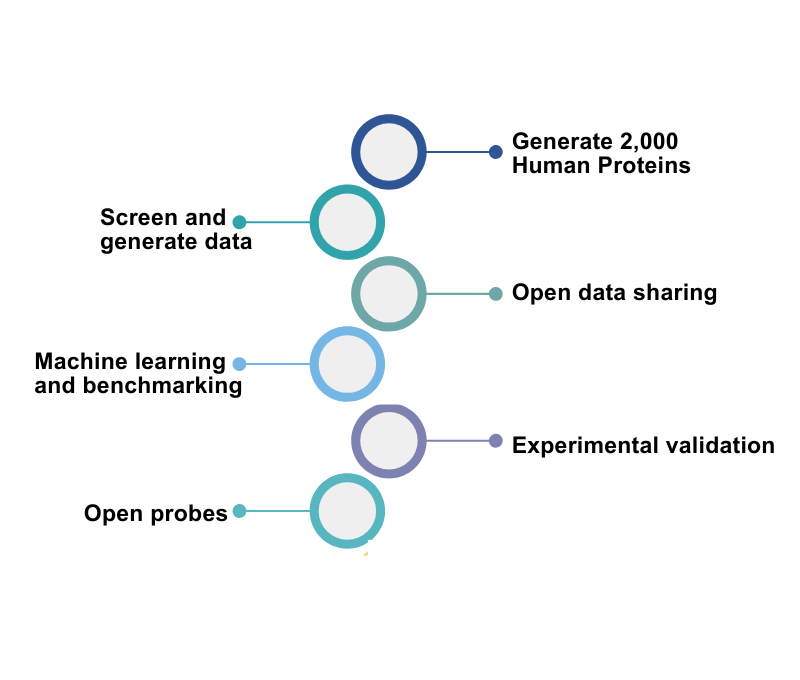
From 2025 onward, the initiative is executing on this now-published vision. It will:
- Generate protein–ligand binding data for over 2,000 diverse human proteins using AS-MS and DEL screening
- Release all data, including negative results, through the ML-ready platform AIRCHECK
- Organize open benchmarking competitions to improve predictive models
- Maintain consistent standards across globally distributed protein production and assay hubs
By 2030, the initiative aims to deliver thousands of experimentally validated small-molecule hits and the computational tools to predict many more.
How to Get Involved
Whether you're a structural biologist, chemoinformatician, or AI researcher, Target 2035 is structured to support your contributions and amplify your impact.
1. Contribute Proteins
Join the Protein Contribution Network and submit purified, high-quality proteins. These will be screened for ligands using advanced platforms. Contributors retain the right to pursue any resulting hits.
2. Participate in Open Benchmarking Challenges
Target 2035 provides a unique platform for computational scientists to benchmark hit-finding algorithms in real-world settings, with experimental testing of model predictions.
The initiative partners with leading open science efforts to design and host benchmarking challenges, including:
- CACHE: Critical Assessment of Computational Hit-Finding Experiments
- CASP: Critical Assessment of protein Structure Prediction
- Target 2035 DREAM Challenges focused on protein–ligand binding predictions
3. Join MAINFRAME
MAINFRAME is a new international network of machine learning researchers, computational chemists, and data scientists. As part of MAINFRAME, you will have access to curated datasets, experimental feedback, and a collaborative environment to test and benchmark your models.