This probe is available at Cayman Chemicals and Sigma.
Click here to obtain the control.
| Probe | Negative control | |
 |
|  |
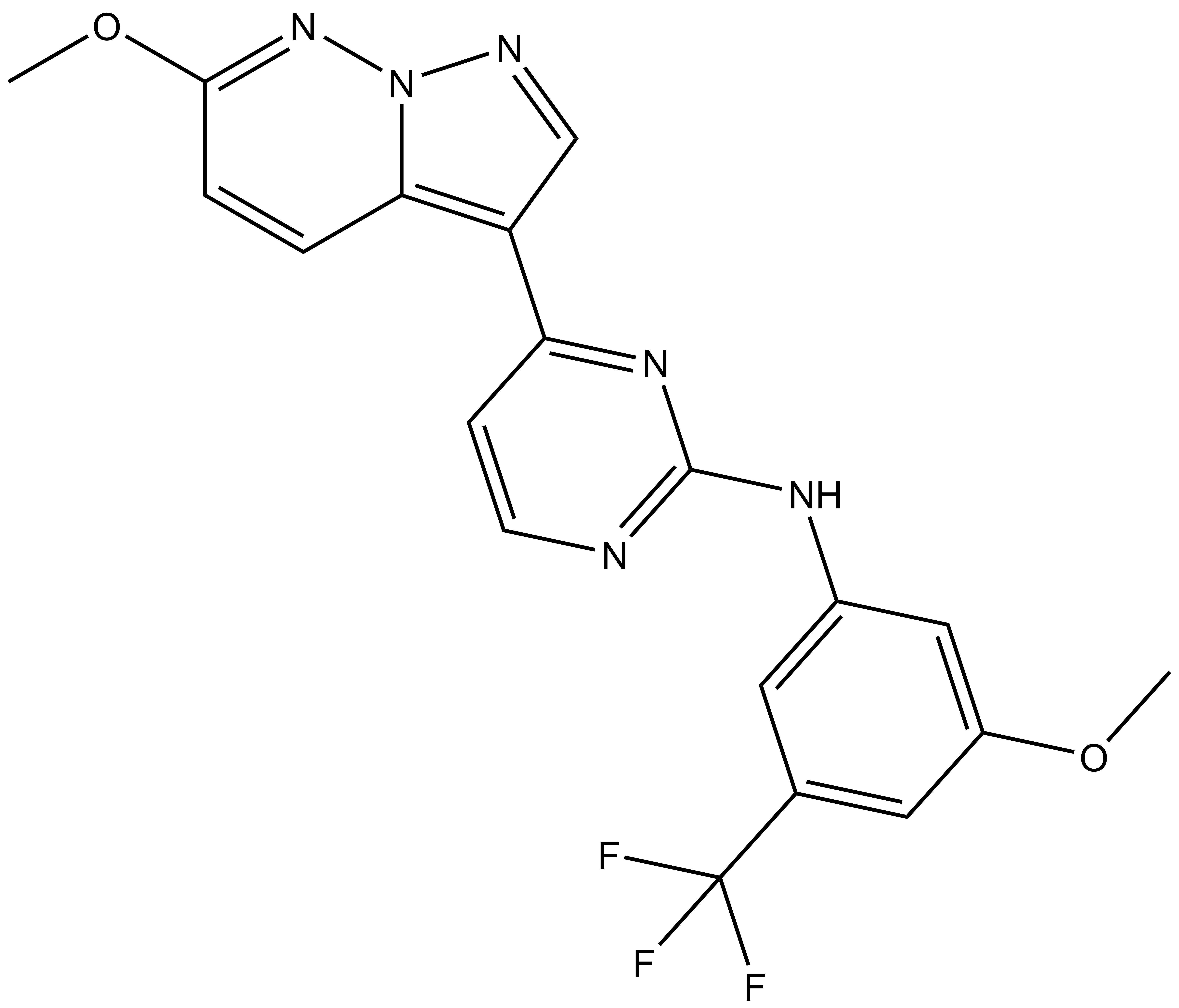
SGC-CLK-1 |
| SGC-CLK-1N |
In collaboration with scientists at Luceome Biotechnologies the SGC has developed a chemical probe for the kinases CLK1, CLK2, and CLK4 and a corresponding closely related negative control compound.
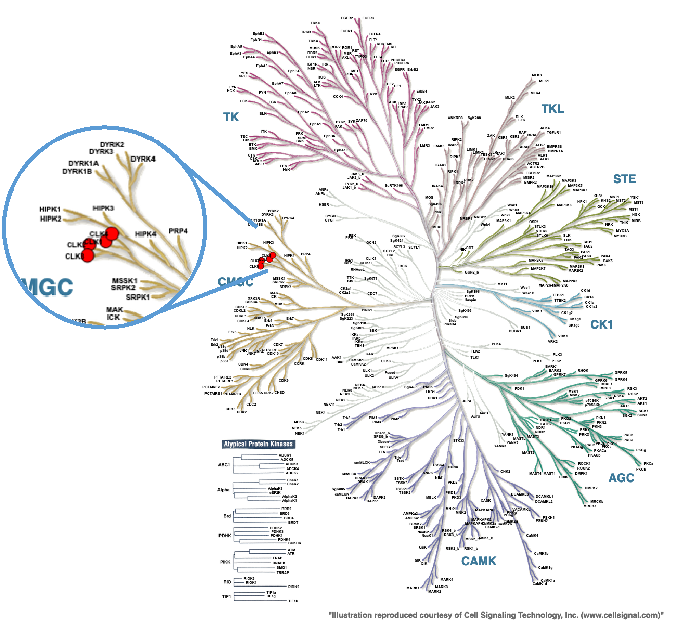
Figure 1: Phylogenetic kinase tree with CLK1, CLK2, CLK3, and CLK4 highlighted with red circles. Illustration is reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com).
The four members of the CDC Like Kinases (CLKs) family (CLK1, CLK2, CLK3, and CLK4) are found in the CMGC branch of the kinome. They are central to RNA-splicing events in cells. CLKs play a role in pre-mRNA splicing is by regulating serine-arginine-rich (SR) proteins. When the SR proteins are phosphorylated by CLKs they relocate to the spliceosome and interact with pre-mRNA to facilitate exon recognition in the splicing machinery. Cancer can gain a survival advantage when the splicing machinery is “hijacked”, so splicing machinery proteins are often mutated or overexpressed in cancers. CLK1, CLK2, and CLK4 for example are overexpressed in renal cancer, breast cancer, colorectal cancer, liver cancer, and glioblastoma. One report suggests a potential vulnerability to CLK inhibitors in cancers with MYC amplification. These findings suggest that CLK inhibitors may be useful for the treatment of some cancers. CLK chemical probes will also help tease out details of the splicing process.
Biological activity summary:
| Probe |
 |
SGC-CLK-1 |
| Physical and chemical properties for SGC-CLK-1 | |
| Molecular weight | 416.4 |
| Molecular formula | C19H15F3N6O2 |
| IUPAC name | N-(3-methoxy-5-(trifluoromethyl)phenyl)-4-(6-methoxypyrazolo[1,5-b]pyridazin-3-yl)pyrimidin-2-amine |
| MollogP | 3.42 |
| PSA | 86.46 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 9 |
| No. of hydrogen bond donors | 1 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10mM) are stable at -20 °C |
| Dissolution | Soluble in DMSO up to 10 mM. |
| Negative control |
 |
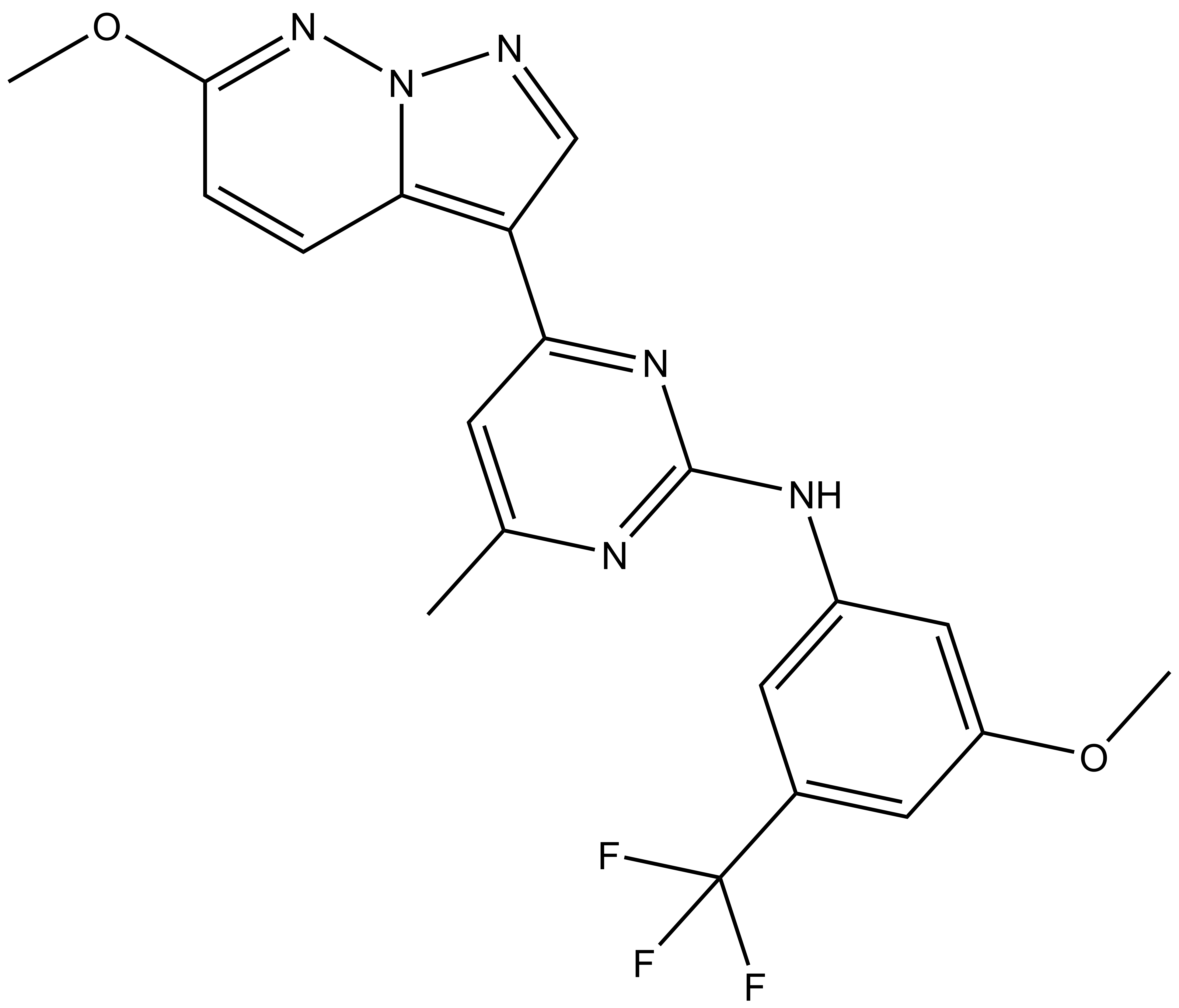
SGC-CLK-1N |
| Physical and chemical properties for SGC-CLK-1N | |
| Molecular weight | 430.4 |
| Molecular formula | C20H17F3N6O2 |
| IUPAC name | N-(3-methoxy-5-(trifluoromethyl)phenyl)-4-(6-methoxypyrazolo[1,5-b]pyridazin-3-yl)-6-methylpyrimidin-2-amine |
| MollogP | 3.81 |
| PSA | 86.46 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 9 |
| No. of hydrogen bond donors | 1 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10mM) are stable at -20 °C |
| Dissolution | Soluble in DMSO |
SMILES:
SGC-CLK-1: COC1=NN2N=CC(C3=NC(NC4=CC(C(F)(F)F)=CC(OC)=C4)=NC=C3)=C2C=C1
SGC-CLK-1N: COC1=NN2N=CC(C3=NC(NC4=CC(C(F)(F)F)=CC(OC)=C4)=NC(C)=C3)=C2C=C1
InChI:
SGC-CLK-1: InChI=1S/C19H15F3N6O2/c1-29-13-8-11(19(20,21)22)7-12(9-13)25-18-23-6-5-15(26-18)14-10-24-28-16(14)3-4-17(27-28)30-2/h3-10H,1-2H3,(H,23,25,26)
SGC-CLK-1N: InChI=1S/C20H17F3N6O2/c1-11-6-16(15-10-24-29-17(15)4-5-18(28-29)31-3)27-19(25-11)26-13-7-12(20(21,22)23)8-14(9-13)30-2/h4-10H,1-3H3,(H,25,26,27)
InChIKey:
SGC-CLK-1: GJYVLTPTDBQQCY-UHFFFAOYSA-N
SGC-CLK-1N: BJVQXSHZMUFJBQ-UHFFFAOYSA-N
SGC-CLK-1 was profiled in the KINOMEscan assay against of 403 wild-type kinases at 1 µM. Only 6 kinases showed PoC < 35 giving an S(35) at 1 µM =0.02. Potential off-targets were tested in the nanoBRET target engagement assay where possible and the data are shown in the table.
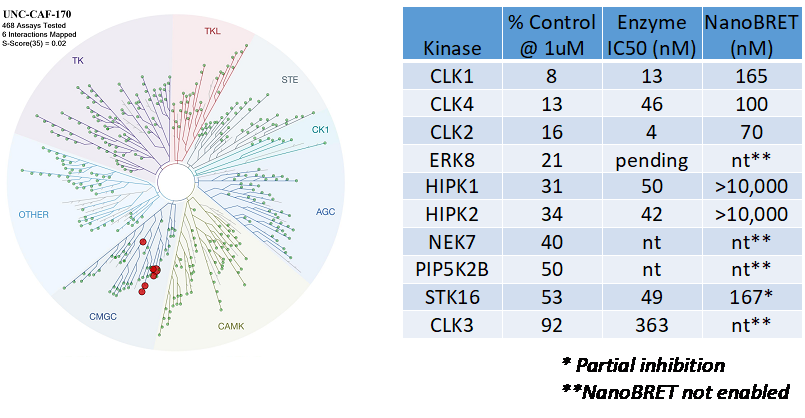
Figure 1: SGC-CLK-1 was profiled in the KINOMEscan assay against of 403 wild-type kinases at 1 µM.
SGC-CLK-1N was also tested in the Discoverx panel and no kinases had PoC<45. The negative control was sent to Eurofins for testing in enzyme assays for CLKs and typical CLK inhibitor off targets. These results are in the table below.
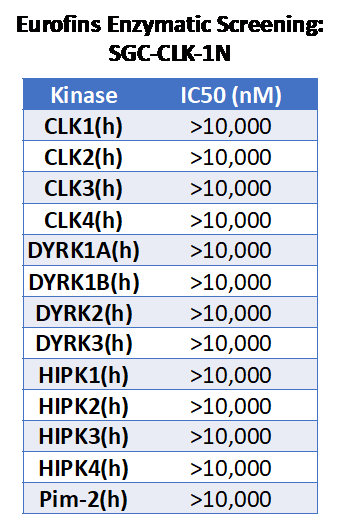
Figure 2: Eurofins enzymatic screening results for SGC-CLK-1.
A Nanobret assay was utilised to assess the binding affinity of SGC-CLK-1 to CLK1, CLK2 and CLK4 (Figure 1). The negative control shows no binding affinity.
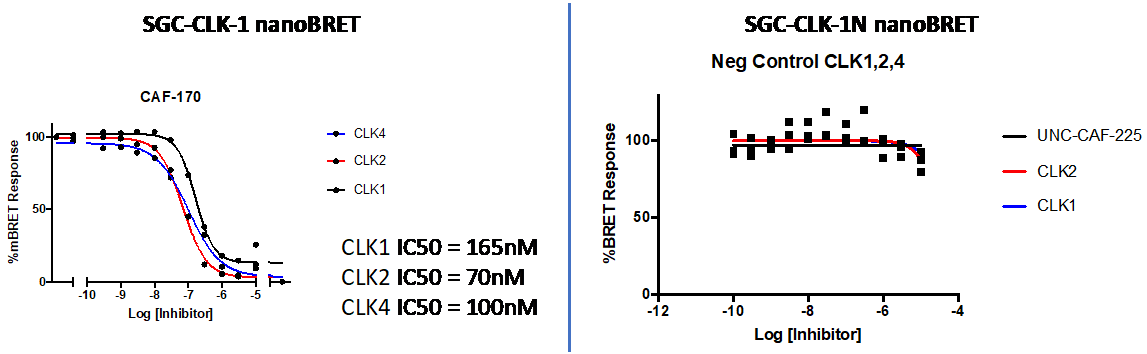
Figure 1: SGC-CLK-1 was profiled in the Nanobret assay.
1. Araki, S., et al., Inhibitors of CLK protein kinases suppress cell growth and induce apoptosis by modulating pre-mRNA splicing. PLoS One, 2015. 10(1): p. e0116929.DOI: 10.1371/journal.pone.0116929. https://www.ncbi.nlm.nih.gov/pubmed/25581376
2. Bullock, A.N., et al., Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation. Structure, 2009. 17(3): p. 352-62.DOI: 10.1016/j.str.2008.12.023. https://www.ncbi.nlm.nih.gov/pubmed/19278650
3. Corkery, D.P., et al., Connecting the speckles: Splicing kinases and their role in tumorigenesis and treatment response. Nucleus, 2015. 6(4): p. 279-88.DOI: 10.1080/19491034.2015.1062194. https://www.ncbi.nlm.nih.gov/pubmed/26098145
4. Funnell, T., et al., CLK-dependent exon recognition and conjoined gene formation revealed with a novel small molecule inhibitor. Nat Commun, 2017. 8(1): p. 7.DOI: 10.1038/s41467-016-0008-7. https://www.ncbi.nlm.nih.gov/pubmed/28232751
5. Iwai, K., et al., Anti-tumor efficacy of a novel CLK inhibitor via targeting RNA splicing and MYC-dependent vulnerability. EMBO Mol Med, 2018. 10(6).DOI: 10.15252/emmm.201708289. https://www.ncbi.nlm.nih.gov/pubmed/29769258
6. jain, P., et al., Human CDC2-like kinase 1 (CLK1): a novel target for Alzheimer's disease. Curr Drug Targets, 2014. 15(5): p. 539-50.DOI: 10.2174/1389450115666140226112321. https://www.ncbi.nlm.nih.gov/pubmed/24568585
7. Koedoot, E., et al., Splicing regulatory factors in breast cancer hallmarks and disease progression. Oncotarget, 2019. 10(57): p. 6021-6037.DOI: 10.18632/oncotarget.27215. https://www.ncbi.nlm.nih.gov/pubmed/31666932
8. Sako, Y., et al., Development of an orally available inhibitor of CLK1 for skipping a mutated dystrophin exon in Duchenne muscular dystrophy. Sci Rep, 2017. 7: p. 46126.DOI: 10.1038/srep46126. https://www.ncbi.nlm.nih.gov/pubmed/28555643