This probe (hydrochloride) is available from Cayman Chemical, Sigma and Tocris.
| Probe | Negative control | |
 |
|  |
TP-238 |
| TP-422 |
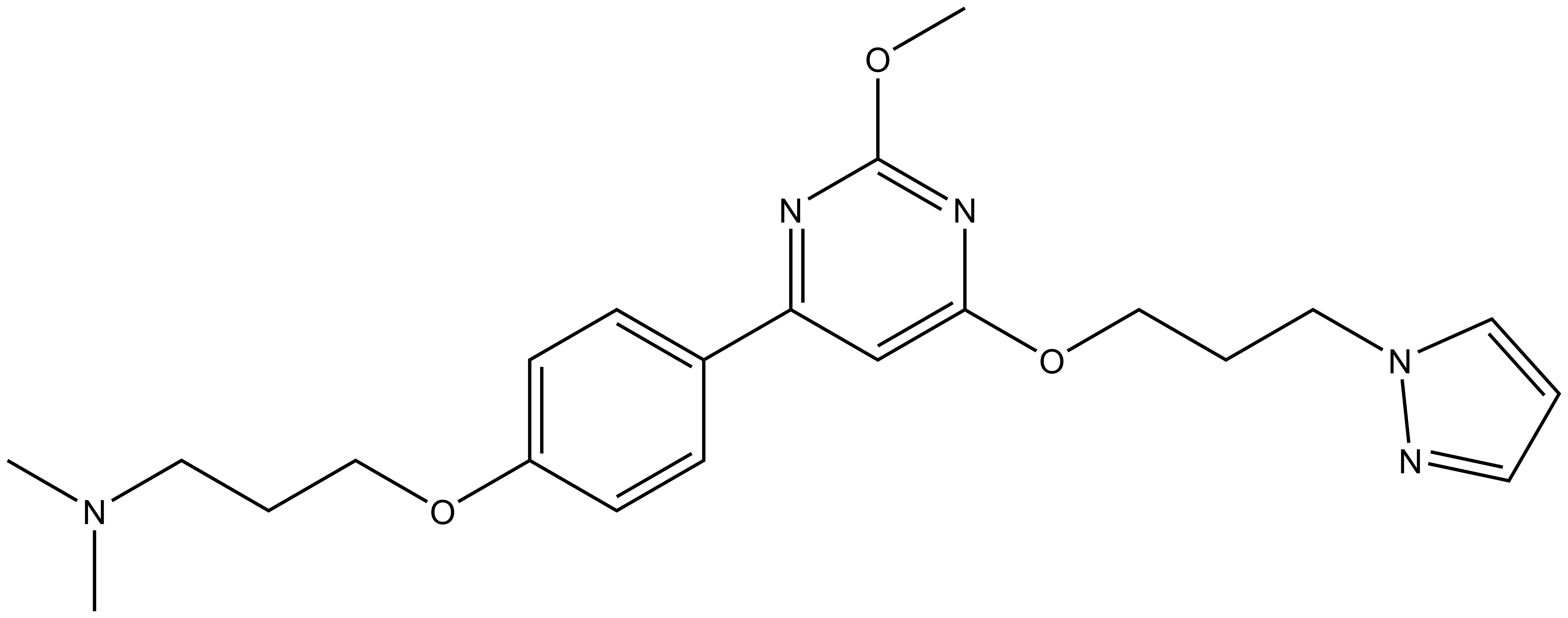
CECR2 (cat eye syndrome chromosome region, candidate 2) gene is predominantly expressed in the nervous system and involved in neurulation. It is located in the segment of chromosome 22q11.2. Multiplication of this segment lead to rare genetic disorder called cat eye syndrome characterized by multiple congenital defects (1). CECR2 has also been implicated in regulation of DNA damage response (2)
Bromodomain PHD finger transcription factor BPTF/FALZ is a core component of the conserved, multi-subunit nucleosome remodelling factor (NURF) complex. BPTF is essential for neural development and haematopoiesis (3). BPTF is involved in c-MYC chromatin recruitment and transcriptional activity in hematopetic and also cancer stem cells (4).
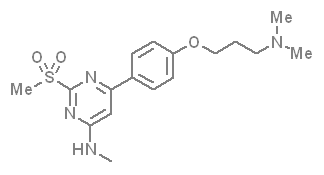
In a collaborative effort Takeda and the SGC have identified and characterised TP-238 as a CECR2/BPTF chemical probe.
TP-238 has on target biochemical activity of 10-30 nM with CECR2 and 100-350 nM with BPTF. Negative control TP-422 is completely inactive against BPTF and CECR2.
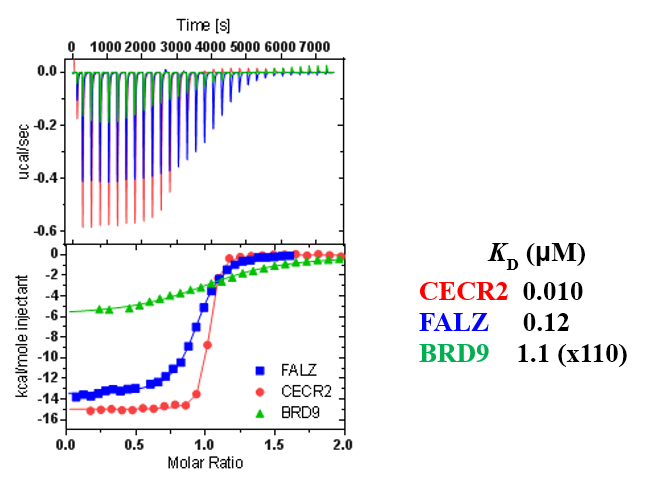
The closest off-target bromodomain inhibition is BRD9 with IC50 of 1.4 µM. TP-238 has been profiled against the panel of 338 kinases and showed no activity at 1 μM.
We recommend that TP-238 and TP-422 be used at no more than 2 µM concentration in cells.
Cell-based NanoBRETTM experiments measured the target engagement with both BPTF and CECR2 with EC50 in the 200-300 nM range.
TP-238 shows an IC50 of 30nM against CECR2 and 350nM against BPTF in an alphascreen assay. ITC shows TP-238 with a KD of 10nM for CECR2 and 120nM for BPTF.
| TP-238 |
 |
Click here to download the SDF file. |
SMILES:
CN(CCCOC1=CC=C(C2=NC(S(C)(=O)=O)=NC(NCCCN3C=CC=N3)=C2)C=C1)C
InChI:
InChI=1S/C22H30N6O3S/c1-27(2)13-6-16-31-19-9-7-18(8-10-19)20-17-21(26-22(25-20)32(3,29)30)23-11-4-14-28-15-5-12-24-28/h5,7-10,12,15,17H,4,6,11,13-14,16H2,1-3H3,(H,23,25,26)
InChIKey:
MSIJJXOWLFOYIN-UHFFFAOYSA-N
| Physical and chemical properties | |
| Molecular weight | 458.21 |
| Molecular formula | C22 H30 N6 O3 S |
| IUPAC name | 1-(3-(6-(4-(3-dimethylamino-propoxy)-phenyl)-2-methylsulfonyl-pyrimidin-4-ylamino)-propyl)-1H-pyrazole |
| clogP | 2.4 |
| PSA | 84.5 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 12 |
| No. of hydrogen bond acceptors | 9 |
| No. of hydrogen bond donors | 1 |
| PAMPA (nm/sec, pH=7.4) | 28 |
| Aqueous solubility (µM, pH= 6.8) | >222 |
| Storage | Store at -20oC |
| Dissolution | Up to 50mM in DMSO |
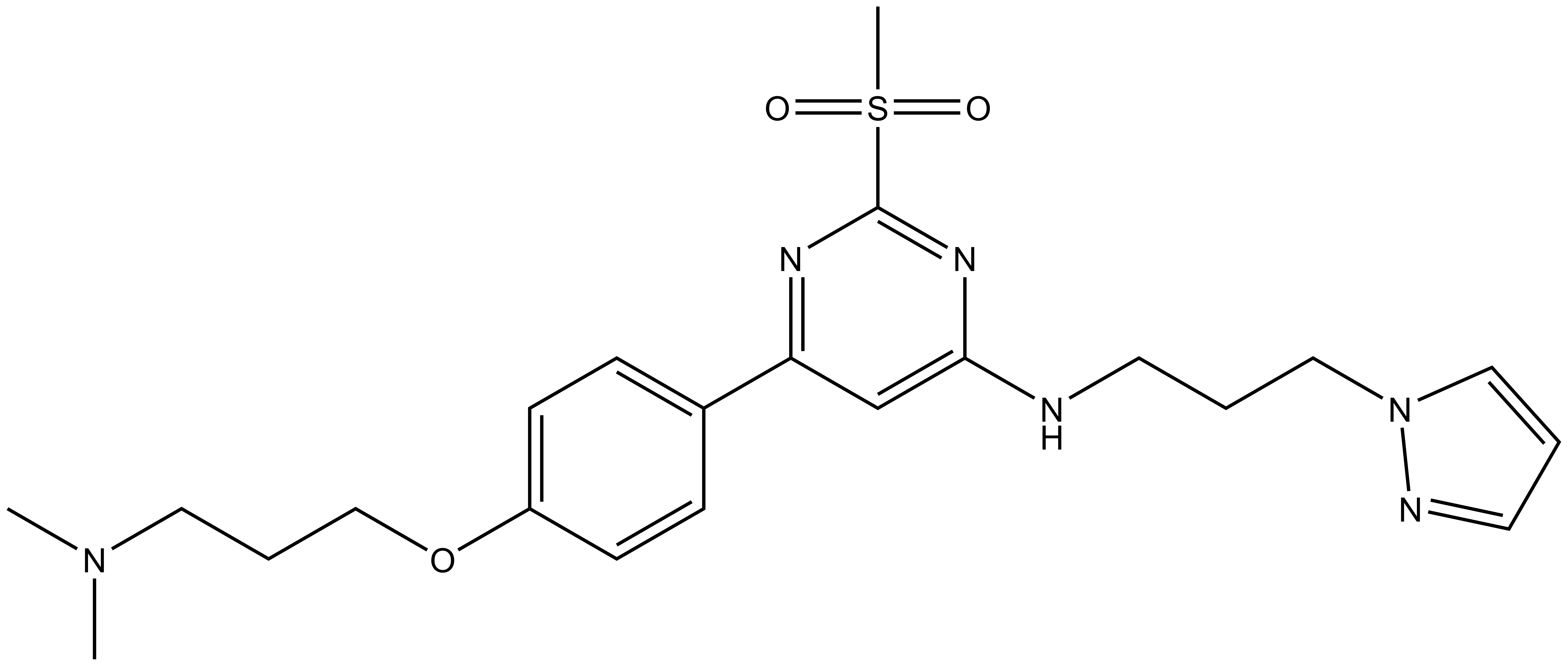
| TP-422 |
 |
Click here to download the SDF file. |
SMILES:
CN(CCCOC1=CC=C(C2=NC(OC)=NC(OCCCN3C=CC=N3)=C2)C=C1)C
InChI:
InChI=1S/C22H29N5O3/c1-26(2)12-5-15-29-19-9-7-18(8-10-19)20-17-21(25-22(24-20)28-3)30-16-6-14-27-13-4-11-23-27/h4,7-11,13,17H,5-6,12,14-16H2,1-3H3
InChIKey:
CVRFBLOQBLKUBC-UHFFFAOYSA-N
| Physical and chemical properties | |
| Molecular weight | 411.22 |
| Molecular formula | C22 H29 N5 O3 |
| IUPAC name | 1-(3-(6-(4-(3-dimethylamino-propoxy)-phenyl)-2-methoxy-pyrimidin-4-yloxy)-propyl)-1H-pyrazole |
| clogP | 3.4 |
| PSA | 59.0 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 12 |
| No. of hydrogen bond acceptors | 7 |
| No. of hydrogen bond donors | 0 |
| PAMPA (nm/sec, pH=7.4) | 283 |
| Aqueous solubility (µM, pH= 6.8) | >267 |
| Storage | Store at -20oC |
| Dissolution | Up to 50mM in DMSO |
A DSF screen against Human bromodomains reveals only one significant off-target; BRD9:
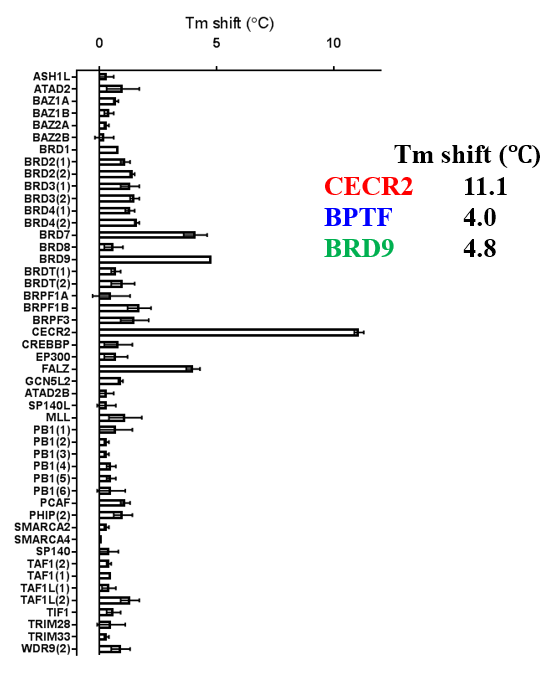
ITC showed good potency and sufficient selectivity against BRD9 to meet SGC probe criteria:
Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Stratagene). Proteins were buffered in 10 mM HEPES pH 7.5, 500 mM NaCl and assayed in a 96-well plate at a final concentration of 2 μM in 20 μL volume. Compounds were added at a final concentration of 10 μM. SYPRO Orange (Molecular Probes) was added as a fluorescence probe at a dilution of 1:1000. Excitation and emission filters for the SYPRO-Orange dye were set to 465 nm and 590 nm, respectively. The temperature was raised with a step of 3 °C per minute from 25 °C to 96 °C and fluorescence readings were taken at each interval.
All bromodomain proteins were prepared according to the published procedures (Filippakopoulos at al, 2012). Assay was performed as described previously (Philpott et al, 2011). All reagents were pre-diluted in 25 mM HEPES, 100 mM NaCl, 0.1 % BSA, pH 7.4 and 0.05 % CHAPS and allowed to equilibrate to room temperature prior to addition to plates. Plates filled with 5 uL of the assay buffer followed by 7 uL of biotinylated peptide [H-YSGRGKacGGKacGLGKacGGAKacRHRK(Biotin)-OH and His-tagged protein to achieve final assay concentrations of 25 nM. Plates were sealed and incubated for a further 60 minutes, before the addition of 8 μl of the mixture of streptavidin-coated donor beads (12.5 μg/ml) and nickel chelate acceptor beads (12.5 μg/ml) under low light conditions. Plates were foil-sealed to protect from light, incubated at room temperature for 60 minutes and read on a PHERAstar FS plate reader (BMG Labtech, Germany) using an AlphaScreen 680 excitation/570 emission filter set.
Experiments were carried out on a VP-ITC microcalorimeter (MicroCal™). All experiments were performed at 15 °C in 25 mM HEPES pH 7.4, 150 mM NaCl, 500 μM TCEP. 50 mM stocks of compound was thawed and diluted in 2 mL of buffer to a final concentration of 10 µM in the ITC cell. The protein titrations were conducted using an initial injection of 2 µl followed by 30 identical injections of 6 µl. The dilution heats were measured on separate experiments and were subtracted from the titration data. Thermodynamic parameters were calculated using ∆G = ∆H - T∆S = -RTlnKB, where ∆G, ∆H and ∆S are the changes in free energy, enthalpy and entropy of binding respectively. In all cases a single binding site model was employed.
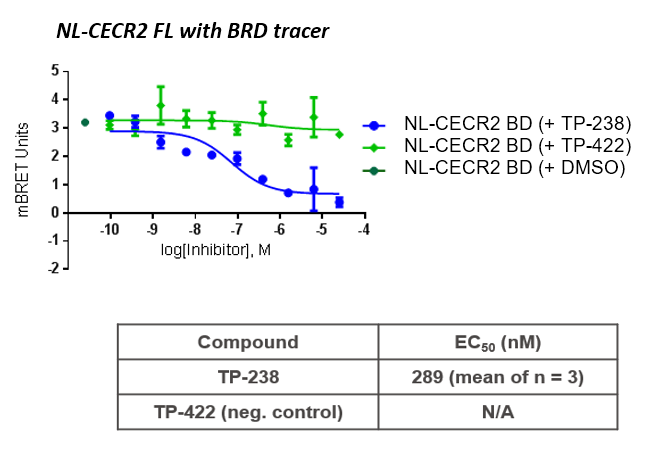
TP-238 and its negative control were tested for target engagement in HEK cells using NanoBRETTM. Significant inhibition by TP-238 against BPTF (n = 3, EC50 = 228 nM) and CECR2 (n = 3, EC50 = 289 nM) whereas the negative control TP-422 exhibited no significant activity at the tested concentrations:
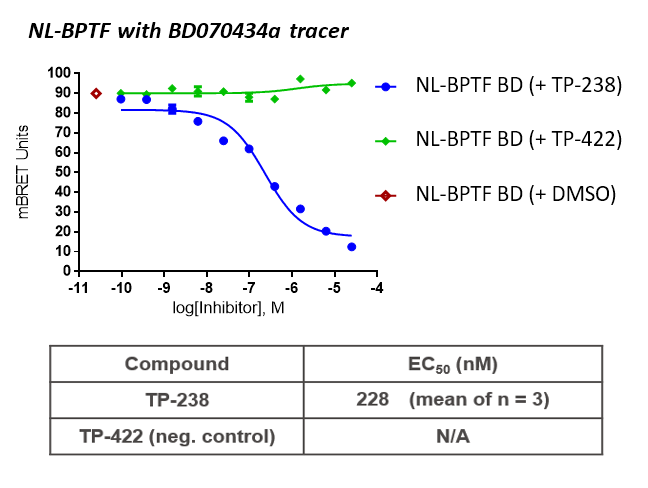
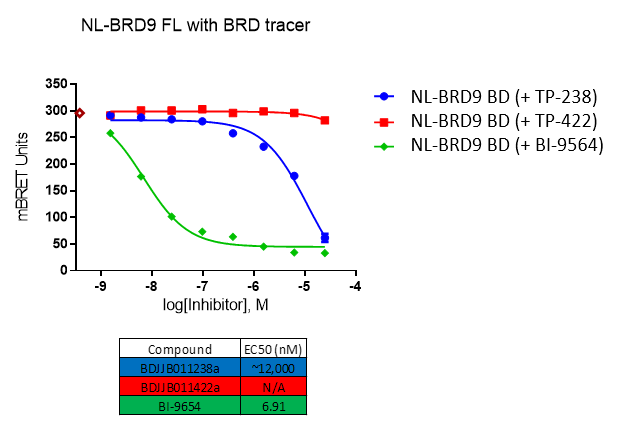
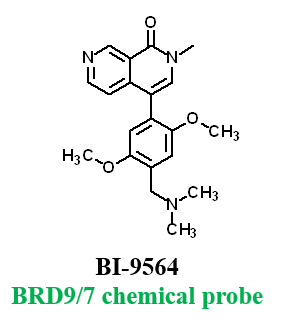
Further, no significant inhibition against BRD9 was found in a NanoBRETTM assay in HEK293 cells:
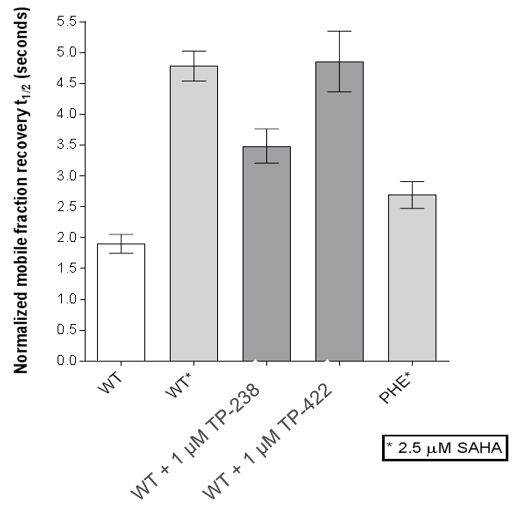
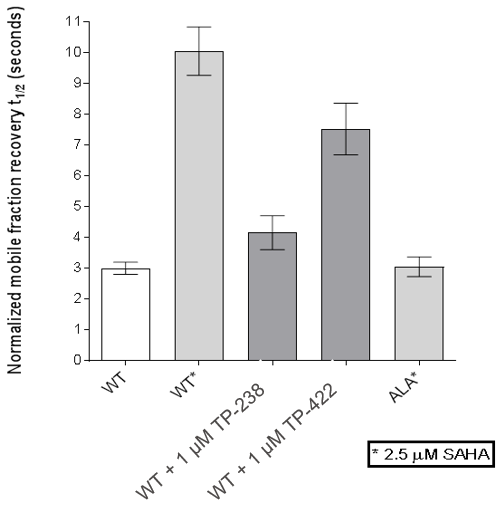
 FRAP assays were also performed which showed significant inhibition by TP-238 against BPTF and CECR2:
FRAP assays were also performed which showed significant inhibition by TP-238 against BPTF and CECR2:
FL-BPTF FRAP:
FL-CECR2 FRAP:
HEK cells were reverse transfected with NL-FL CECR2 or NL-BPTF BD. Cells were treated with a concentration range of TP-238, TP-422 (negative control), with BD070434a tracer (1 µM) for BPTF or BRD-02 tracer (0.5 µM) for CECR2 for 3 hrs before BRET measurements were taken.
For BRD9, the full length protein was used. HEK2983 cells were transfected with NL-BRD9 and treated with tracer (2 µM) and the respective compounds, followed by addition of Nano-Glo substrate and extracellular inhibitor. BRET was determined at 450 and 610nM.
FRAP assays were performed using U2OS cells reverse transfected with GFP-FL FALZ WT or #PHE or GFP-FL CECR2 WT or #ALA for 6-8 hrs. Transfection reagents were replaced with media or 2.5 µM SAHA + incubated O/N. Cells were treated with 1 µM test cpds for 1 hr before imaging. 6 repeat assays were performed for each protein.
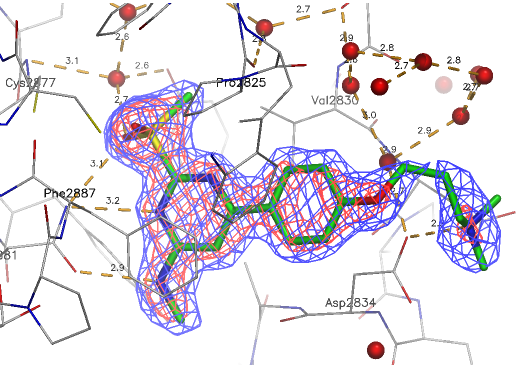
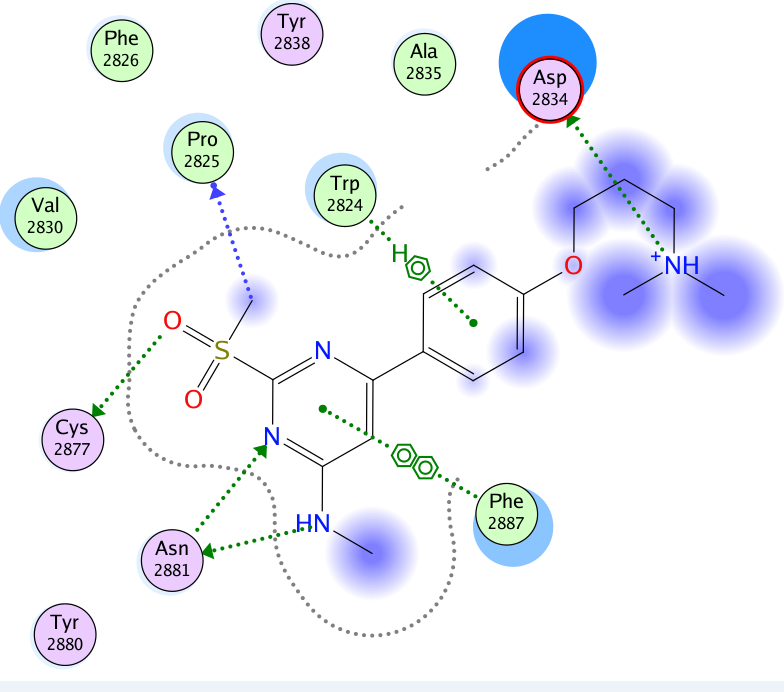
A close homologue to TP-248 was co-crystallised to investigate the possible binding mode of the probe: