This probe is available from Sigma, Cayman Chemical and Tocris.
The control may be requested by clicking here.
| Probe | Negative control | Alternative control | ||
 |
|  |  | |
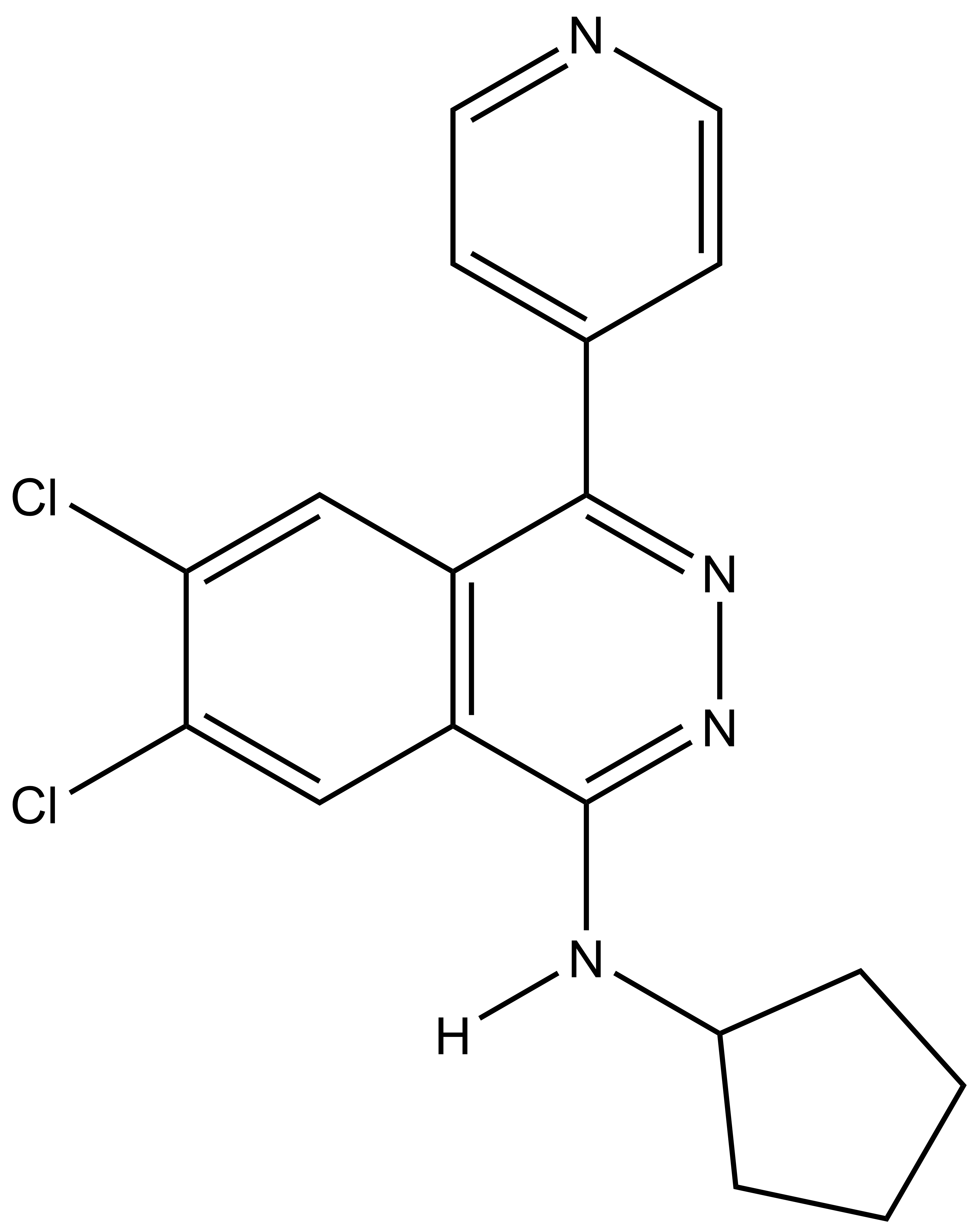
A-196 |
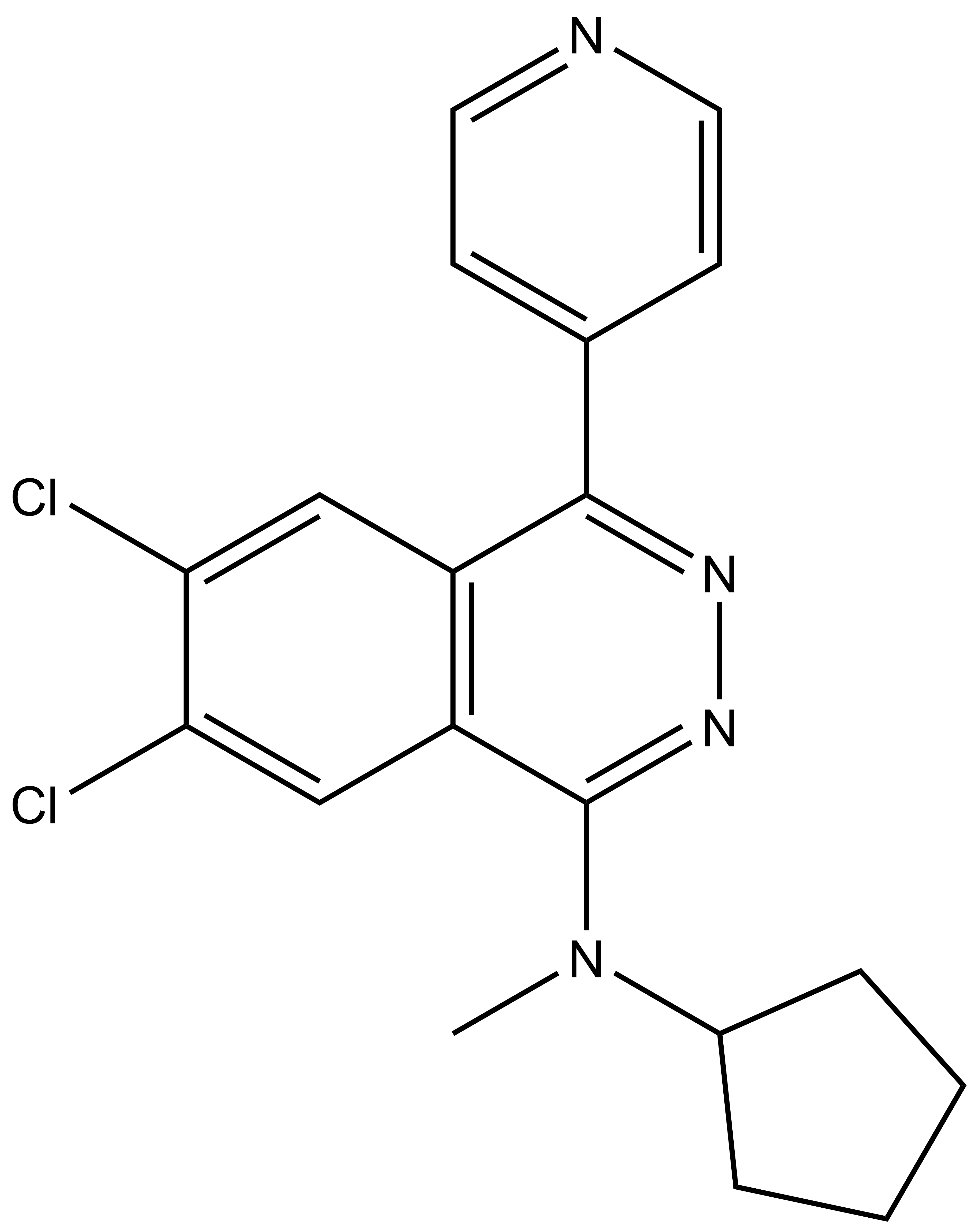
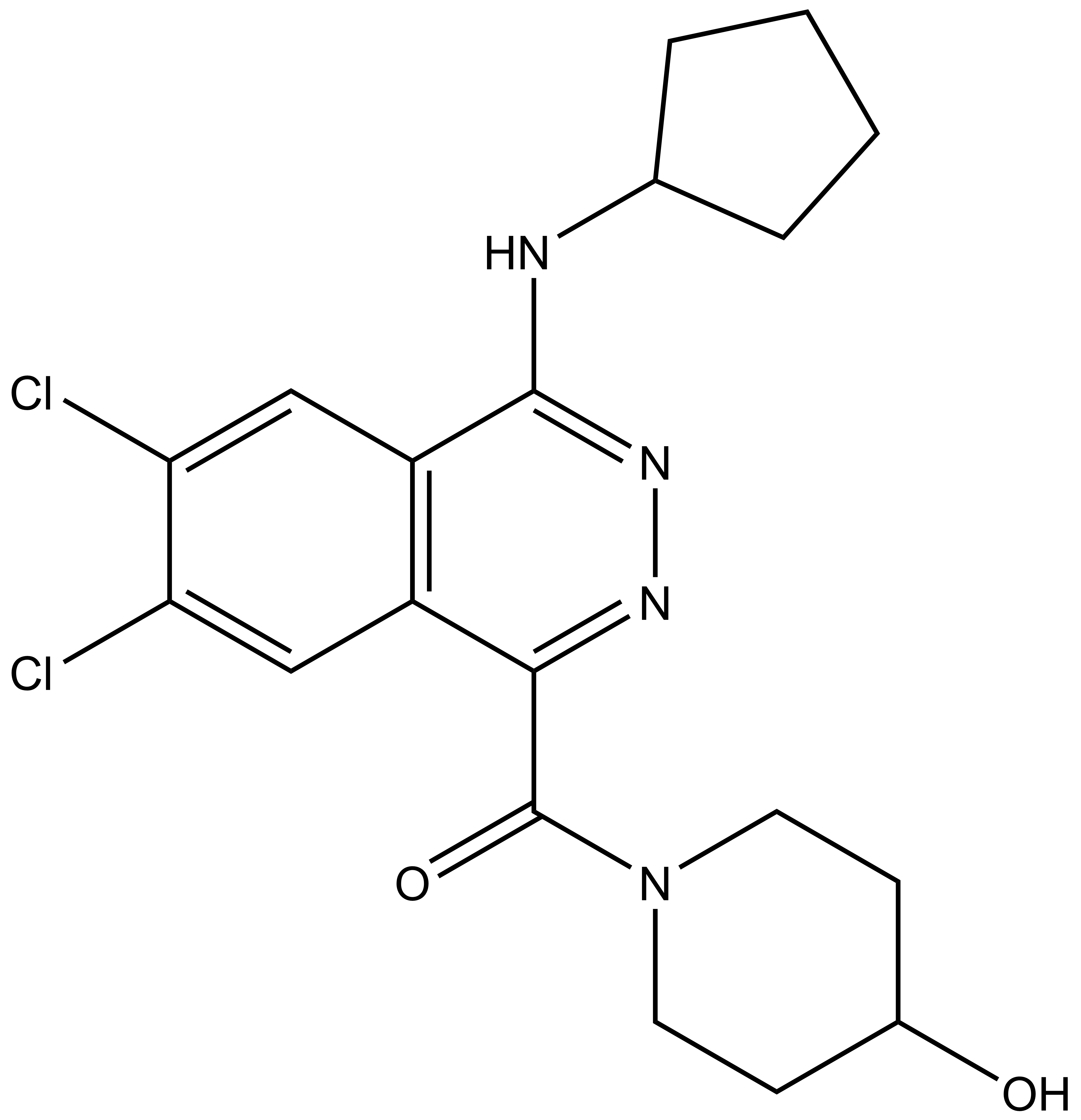
| SGC2043 | A-197 |
Histone H4 lysine 20 methylation is emerging as a crucial modification to ensure genomic integrity both in the absence and presence of genotoxic stress [1]. The majority of histone H4 methylation is detected in the lysine 20 (H4K20) and is evolutionarily conserved from yeast to human [2,3]. Each methylation state results in distinct biology: dimethylated H4K20 (H4K20me2) is involved in DNA replication and DNA damage repair, and trimethylated H4K20 (H4K20me3) results in silenced heterochromatic regions [1]. Loss of histone H4 lysine 20 trimethylation (H4K20me3) is characteristic of human cancer and a potential prognostic marker in many types of cancers [4].
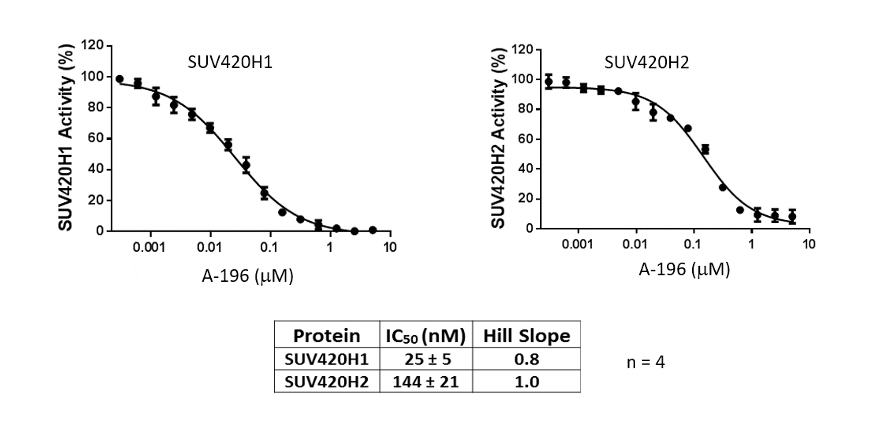
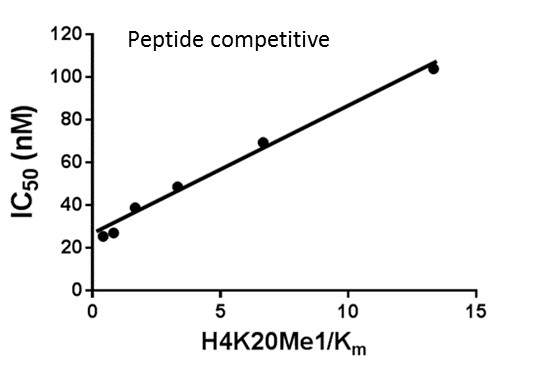
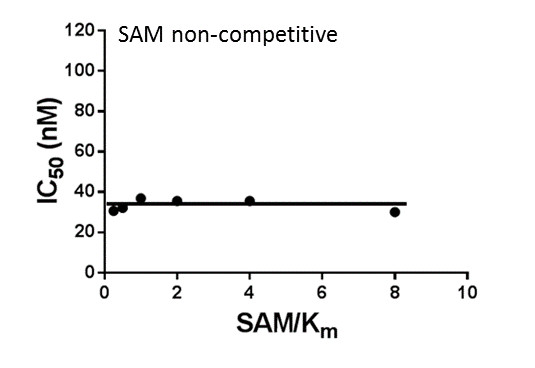
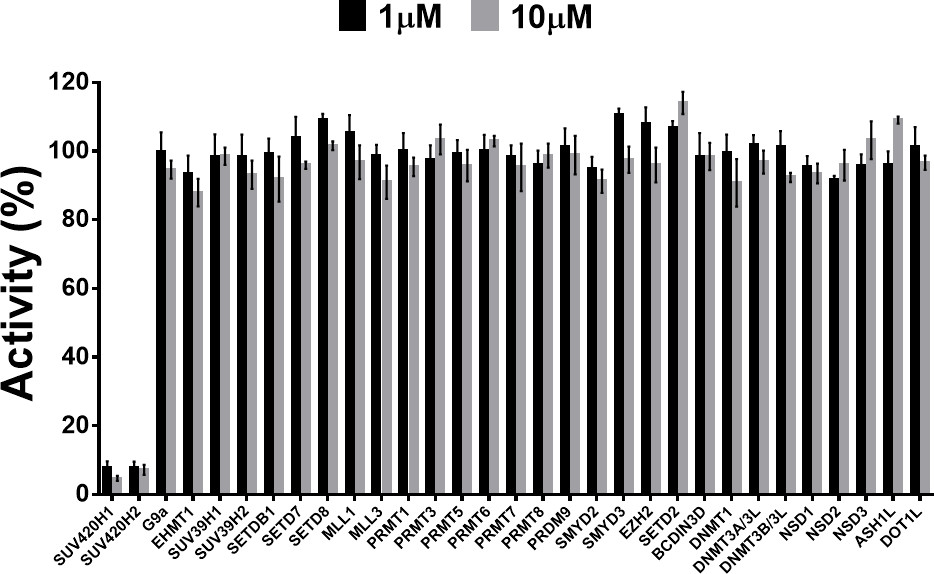
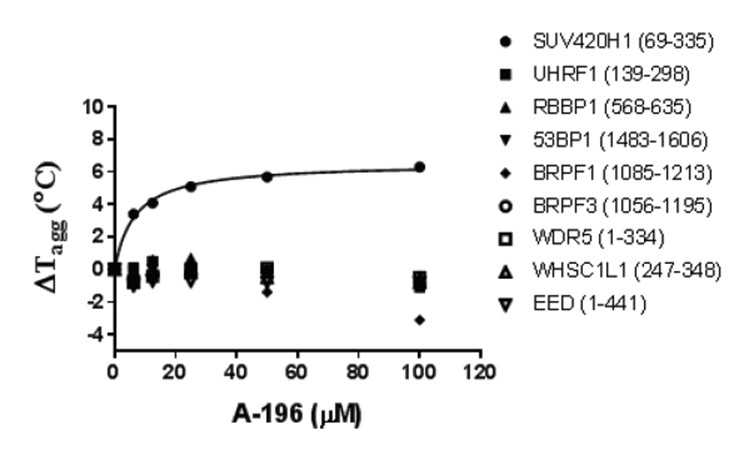
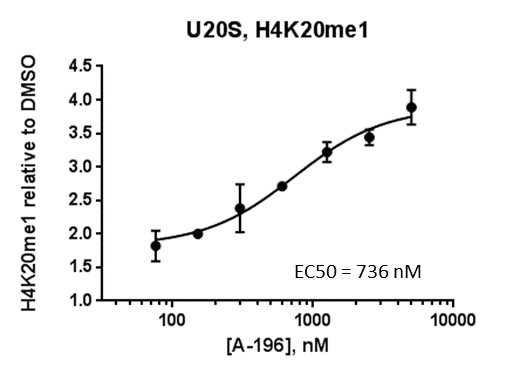
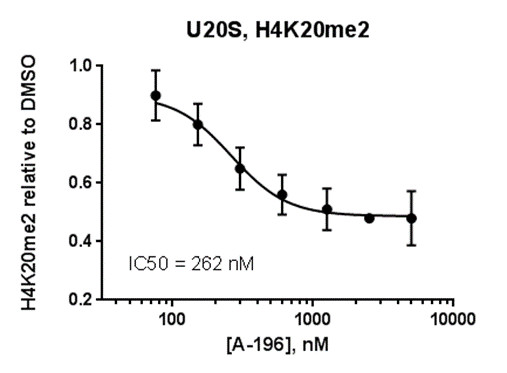
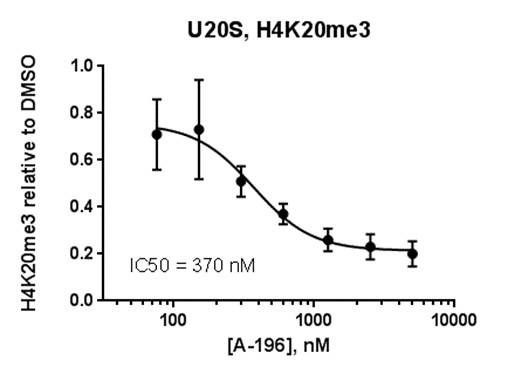
SUV420H1 and SUV420H2 are two highly homologous methyltransferases that di- and tri-methylate 'Lys-20' of histone H4. A collaboration between AbbVie and the SGC has resulted in the discovery of A-196 [5], the first potent and selective chemical probe for SUV420H1 and SUV420H2. The in vitro activity of A-196 includes inhibition of SUV420H1 with IC50 = 25 nM and SUV420H2 with IC50 = 144 nM for methylation of H4K20me and greater than 100-fold selectivity over other histone methyltransferases and non-epigenetic targets. In cell assays, A-196 inhibits the di- and tri-methylation of H4K20me in multiple cell lines with IC50 < 1 µM.
 |  |
| Probe | Negative control | Alternative control | ||
 |
|  |  | |
A-196 |
| SGC2043 | A-197 |
| Physical and chemical properties for A-196 | |
| Molecular weight | 358.1 |
| Molecular formula | C18H16Cl2N4 |
| IUPAC name | 8,9-dichloro-N-cyclopentyl-5-(pyridin-4-yl)-3,4-diaza-bicyclo[4.4.0]deca-1(10),2,4,6,8-pentaen-2-amine |
| MollogP | 5.04 |
| PSA | 42.6 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 3 |
| No. of hydrogen bond acceptors | 3 |
| No. of hydrogen bond donors | 1 |
| Physical and chemical properties for A-197 | |
| Molecular weight | 408.1 |
| Molecular formula | C19H22Cl2N4O2 |
| IUPAC name | (8,9-dichloro-5-cyclopentylamino-3,4-diaza-bicyclo[4.4.0]deca-1(10),2,4,6,8-pentaen-2-yl)-(4-hydroxy-piperidin-1-yl)-methanone |
| MollogP | 3.689 |
| PSA | 65.82 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 4 |
| No. of hydrogen bond acceptors | 5 |
| No. of hydrogen bond donors | 2 |
 |  |  |
[1] Jorgensen S, Schotta G, Sorenson CS. (2013) Histone H4 Lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Research 41: 2797-806.
[2] Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, et al. (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251–1262.
[3] Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, et al. (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell; 119:603–614.
[4] Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, et al. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37:391-400.
[5] Bromberg KD, Mitchell TR, Upadhyay AK, Jakob CG, Jhala MA, et al. (2017 Jan 23) The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity. Nat Chem Biol.