This probe is available from Sigma, Cayman Chemical and Tocris.
The control may be requested by clicking here.
| Probe | Negative control | |
 |
|  |
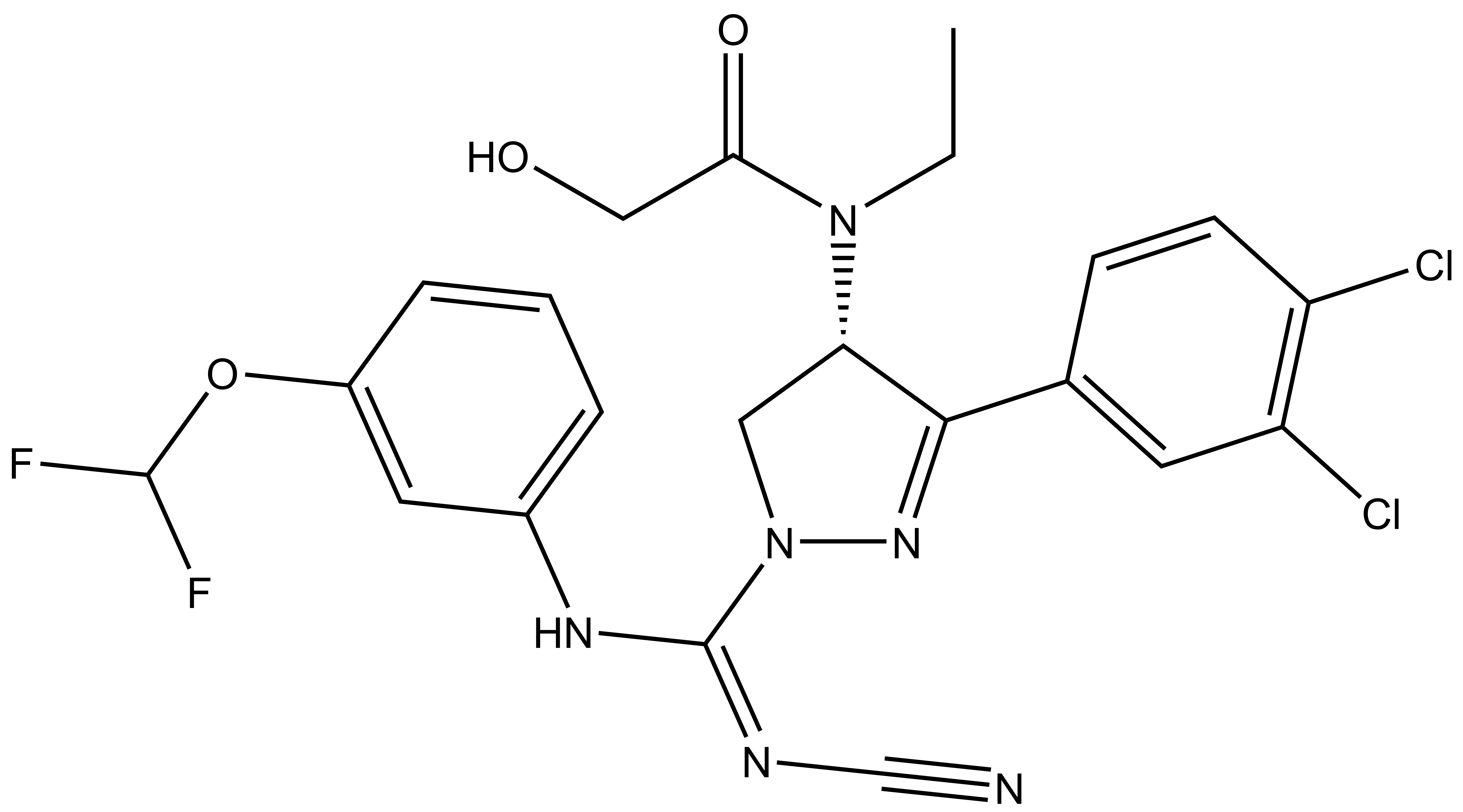
BAY-598 |
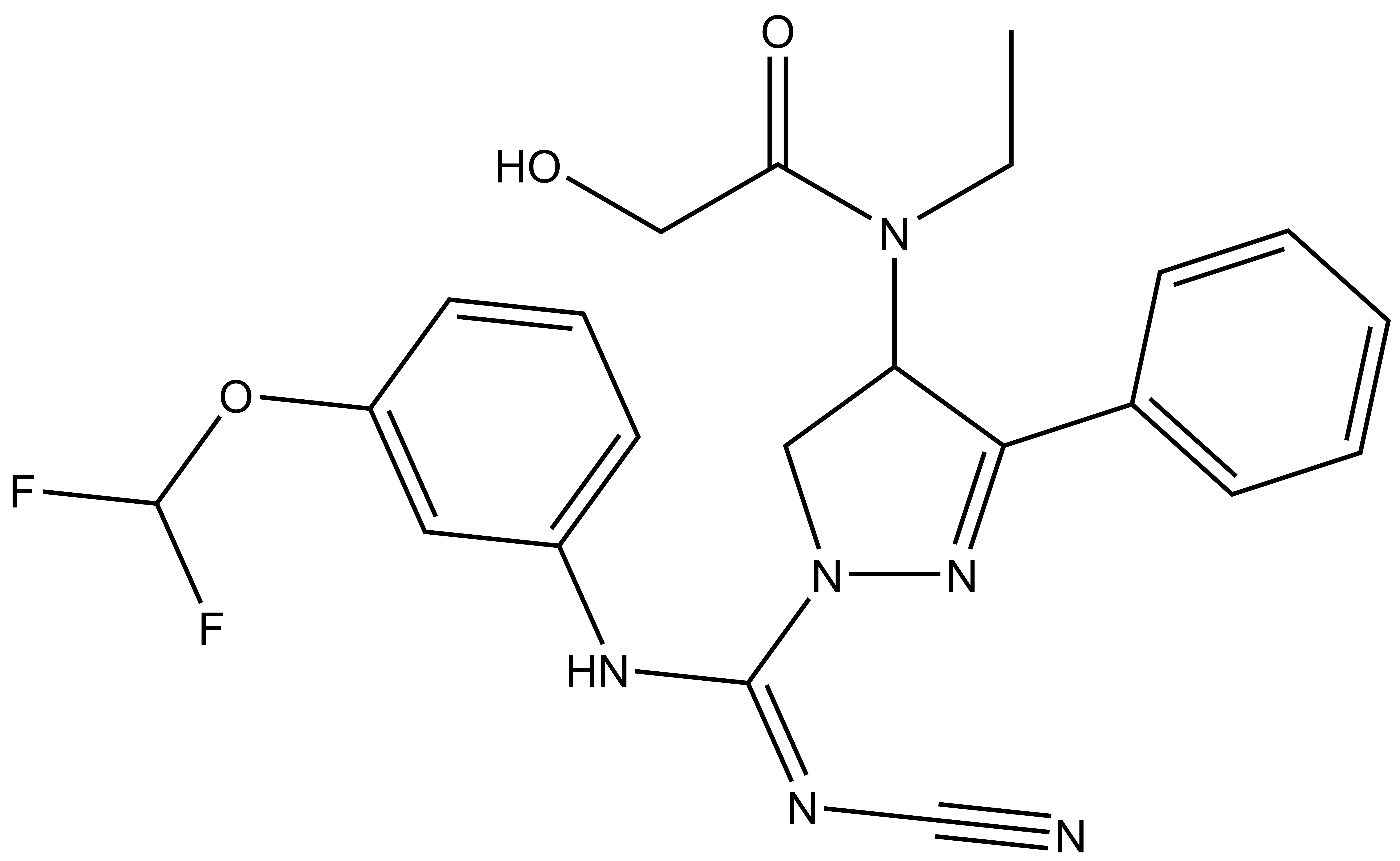
| BAY-369 |
SET and MYND domain-containing protein 2 (SMYD2) is a member of the SMYD family of protein methyltransferases. All five members of this family (SMYD1–5) contain a conserved catalytic SET domain and a zinc-finger MYND motif. SMYD2 methylates both histone and non-histone proteins, including p53/TP53 and RB1 [1-3]. It specifically methylates histone H3 'Lys-4' (H3K4me) and dimethylates histone H3 'Lys-36' (H3K36me2) [1]. It has relatively higher methyltransferase activity on p53/TP53 and monomethylates 'Lys-370' of p53/TP53, leading to decreased DNA-binding activity and subsequent transcriptional regulation activity of p53/TP53. SMYD2 is over-expressed in esophageal squamous primary carcinomas and that over-expression correlates with poor patient survival [2].
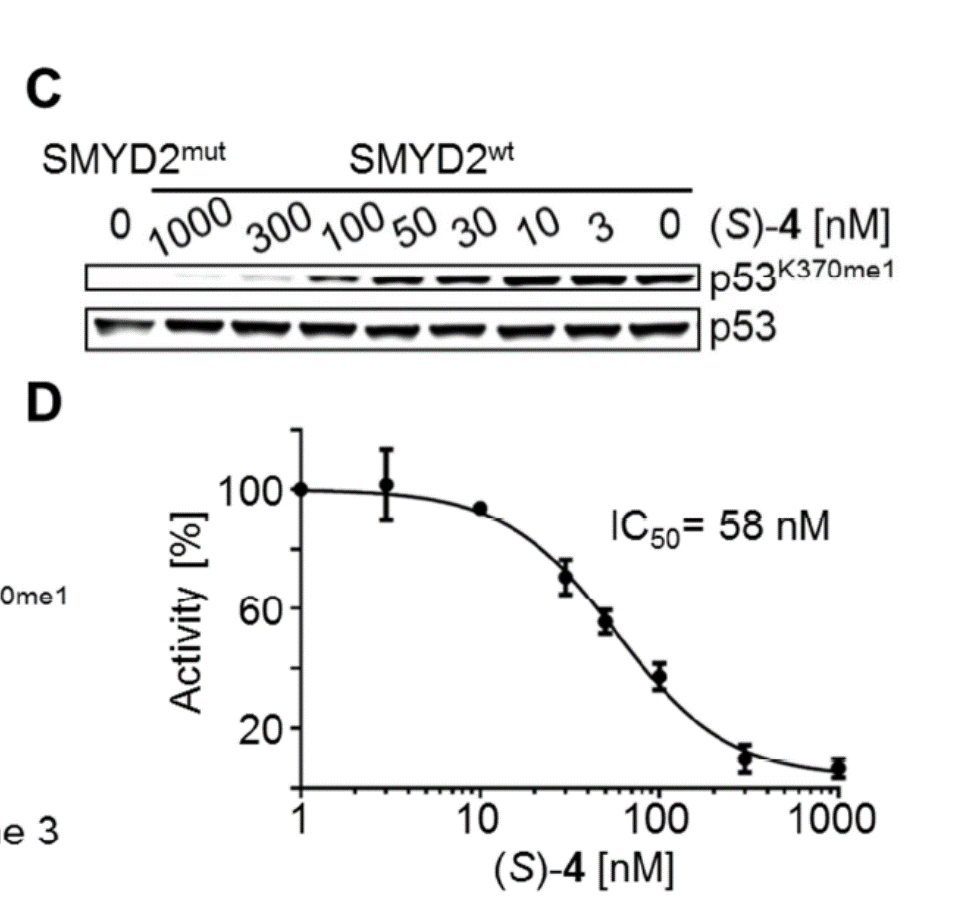
A collaboration between Bayer and the SGC has resulted in the discovery of BAY-598 [4], a potent, peptide-competitive chemical probe for SMYD2. BAY-598 has a unique chemotype relative to the current SMYD2 chemical probe LLY-507 [5] and inhibitors [6,7]. BAY-598 inhibits in vitro methylation of p53K370 with IC50 = 27 nM and has more than 100-fold selectivity over other histone methyltransferases and other non-epigenetic targets. BAY-598 inhibits the methylation of p53K370 in cells with IC50 < 1 µM. (Further to this, BAY-598 has properties that are compatible with in vivo experiments.) A control compound, BAY-369, has also been developed. BAY-369 inhibits the in vitro methylation of p53K370 with IC50 > 70 micromolar.
| Probe | Negative control | |
 |
|  |
BAY-598 |
| BAY-369 |
| Physical and chemical properties for BAY-598 | |
| Molecular weight | 524.1 |
| Molecular formula | C22H20Cl2F2N6O3 |
| MollogP | 3.881 |
| PSA | 86.02 |
| No. of chiral centres | 1 |
| No. of rotatable bonds | 11 |
| No. of hydrogen bond acceptors | 7 |
| No. of hydrogen bond donors | 2 |
| Physical and chemical properties for BAY-369 (racemate) | |
| Molecular weight | 456.2 |
| Molecular formula | C22H22F2N6O3 |
| MollogP | 2.573 |
| PSA | 86.02 |
| No. of chiral centres | 1 |
| No. of rotatable bonds | 11 |
| No. of hydrogen bond acceptors | 7 |
| No. of hydrogen bond donors | 2 |
SMILES:
BAY-598: CCN([C@H]1CN(N=C1C2=CC(Cl)=C(Cl)C=C2)/C(NC3=CC=CC(OC(F)F)=C3)=N\C#N)C(CO)=O
BAY-369: CCN(C(CO)=O)C1CN(N=C1C2=CC=CC=C2)/C(NC3=CC=CC(OC(F)F)=C3)=N\C#N
InChI:
BAY-598: InChI=1S/C22H20Cl2F2N6O3/c1-2-31(19(34)11-33)18-10-32(30-20(18)13-6-7-16(23)17(24)8-13)22(28-12-27)29-14-4-3-5-15(9-14)35-21(25)26/h3-9,18,21,33H,2,10-11H2,1H3,(H,28,29)/t18-/m0/s1
BAY-369: InChI=1S/C22H22F2N6O3/c1-2-29(19(32)13-31)18-12-30(28-20(18)15-7-4-3-5-8-15)22(26-14-25)27-16-9-6-10-17(11-16)33-21(23)24/h3-11,18,21,31H,2,12-13H2,1H3,(H,26,27)
InChIKey:
BAY-598:OTTJIRVZJJGFTK-SFHVURJKSA-N
BAY-369: OEYJKIQNPLLSFW-UHFFFAOYSA-N
BAY-598 is a potent SMYD2 inhibitor and acts via a peptide competitive mechanism of action.
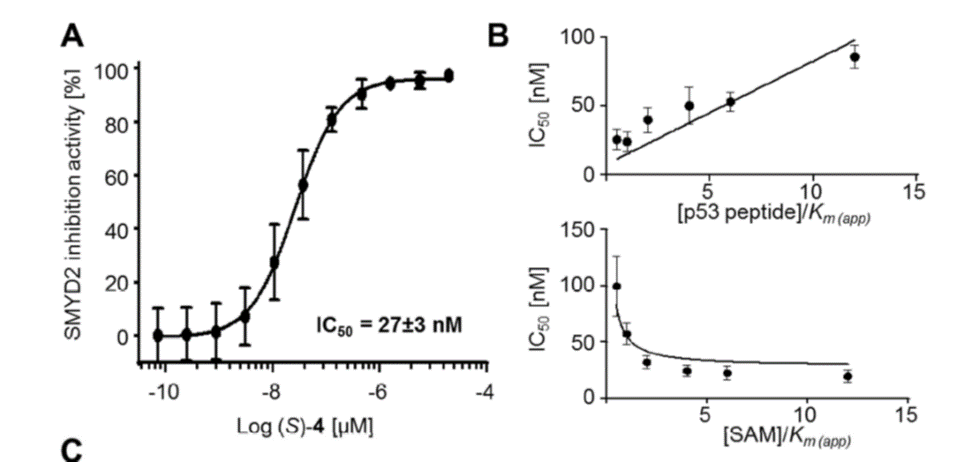
Selectivity
Selectivity of BAY-598 within methyltransferase family
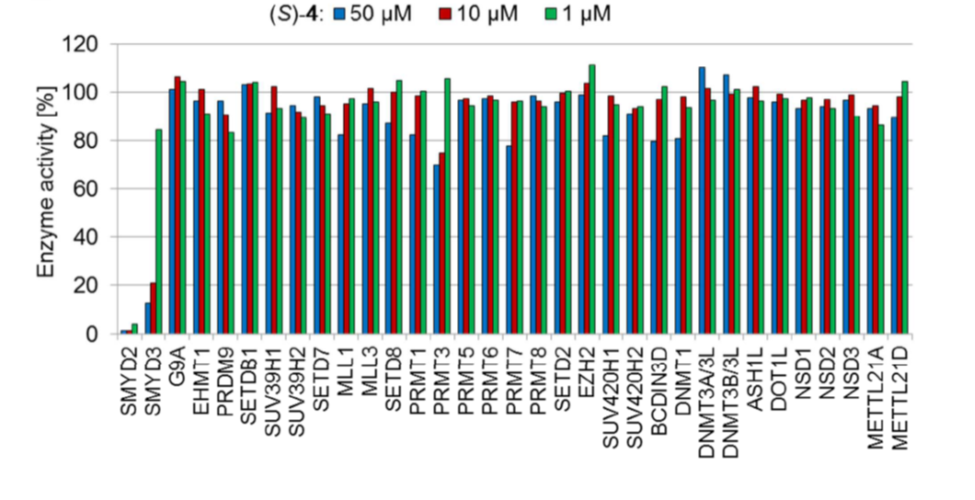
Cellular activity
BAY-598 is a potent inhibitor of p53 K370 methylation in cells
1.Brown MA, Sims RJ 3rd, Gottlieb PD, Tucker PW (2006) Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer 5: 26.
2.Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, et al. (2009) Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis 30: 1139-1146.
3.Cho HS, Hayami S, Toyokawa G, Maejima K, Yamane Y, et al. (2012) RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia 14: 476-486.
4.Eggert E, Hillig RC, Köhr S, Stöckigt D, Weiske J, Barak N,, et al. (2016) Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in vivo Probe (BAY-598) for the Protein Lysine Methyltransferase SMYD2 J Med Chem
5.Nguyen H, Allali-Hassani A, Antonysamy S, Chang S, Chen LH, et al. (2015) LLY-507, a Cell-Active, Potent and Selective Inhibitor of Protein Lysine Methyltransferase SMYD2. 290: 13641-53.
6.Ferguson AD, Larsen NA, Howard T, Pollard H, Green I, et al. (2011) Structural Basis of Substrate Methylation and Inhibition of SMYD2. Structure 19: 1262-73.
7.Sweis RF, Wang Z, Algire M, Arrowsmith CH, Brown PJ, et al. (2015) Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2. ACS Med Chem Lett. 6: 695-700.
Main features