The probe GSK484 (hydrochloride) is available from Cayman Chemical and Sigma.
The negative control GSK106 (hydrochloride) is available from Cayman Chemical.
| Probe | Negative control | |
 |
|  |
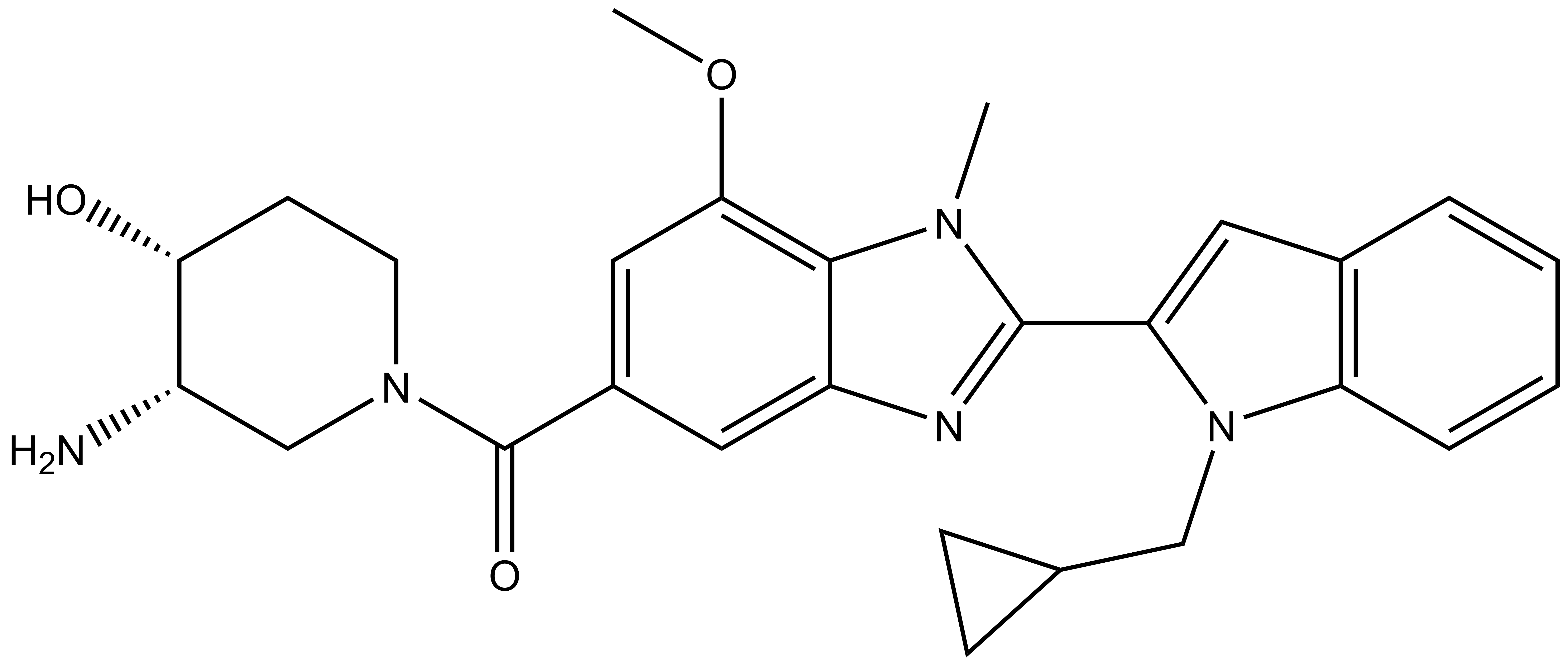
GSK484 |
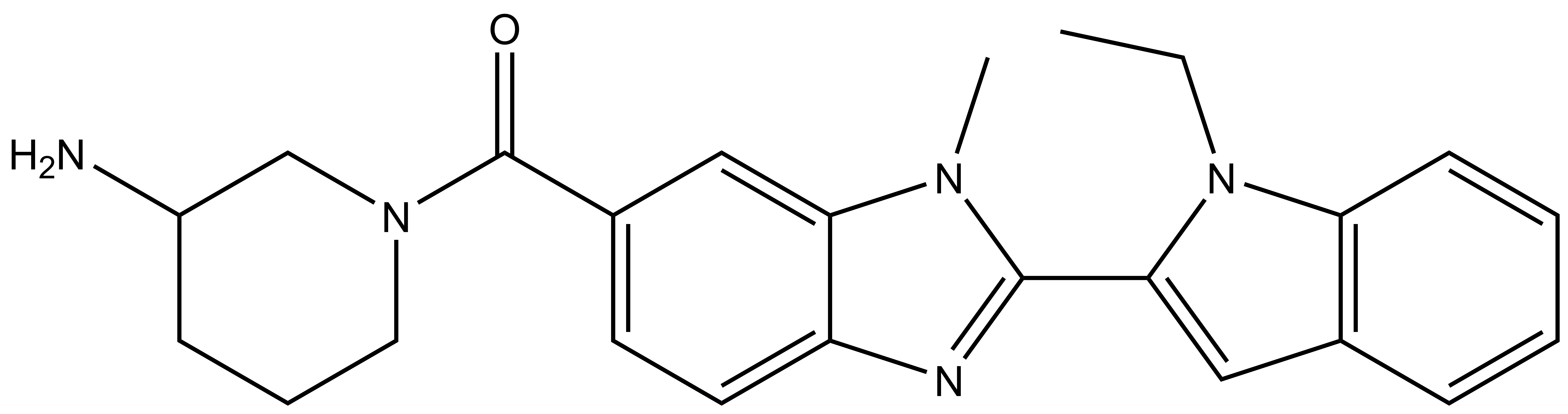
| GSK106 |
PAD4 is a calcium-dependent enzyme which catalyses the transformation of protein arginine residues into citrulline, with the release of ammonia. PAD4-dependent citrullination/deimination of histones plays a key role in the histone code and is predicted to manifest with wide-ranging transcriptional and structural functions, including recently-discovered roles in the regulation of stem cell maintenance (1). In addition to a growing rationale in oncology, PAD4 has strong associations with multiple immune and inflammatory processes. For example, in rheumatoid arthritis the enzyme citrullinates joint proteins to break tolerance and provoke autoimmunity, with antibodies against these citrullinated epitopes (and against PAD4 itself) representing a diagnostic hallmark of the disease. In addition, PAD4 is known to promote profound chromatin decondensation during the innate immune response to infection in neutrophils by mediating formation of neutrophil extracellular traps (NETs). This is an enigmatic and exciting field where initially proposed roles for NETs in trapping pathogens for host-defence purposes (2) have been extended to demonstrate that unrestrained NETosis may be crucial for pathological deep venous thrombosis (3) ischemia/reperfusion injury (4), systemic lupus erythematosis (5), small vessel vasculitis (6) and also in rheumatoid arthritis (7).
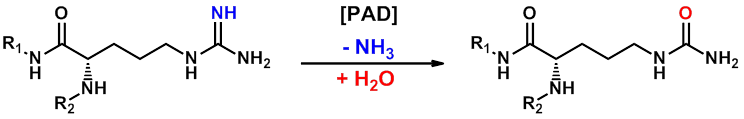
GlaxoSmithKline has developed a PAD4-specific probe (8), namely GSK484 and has made it available as part of the SGC epigenetics initiative. GSK484 potently binds to the low-calcium form of PAD4 in a reversible manner (IC50 of 50 nM) and appears to be competitive with substrate. GSK106 is a related control molecule (IC50 > 100 µM) which offers important confirmation of PAD4-specific effects. Detailed crystallography work with this compound series has additionally demonstrated binding to a new conformation of the PAD4 active site where key residues are re-ordered to form a β-hairpin. GSK484’s selectivity for PAD4 over PAD1-3 was shown in cells and also confirmed with recombinant enzymes. This probe is an inhibitor of cellular citrullination in primary neutrophils, and further phenotypic profiling has confirmed its ability to inhibit NET formation in both mouse and human neutrophils. GSK484 exhibits favourable pharmacokinetic profiles, with low-moderate clearance, and good volume of distribution and half-life in mouse and rat, and has suitable a PK profile for use as a potential in vivo tool.
| Probe | Negative control | |
 |
|  |
GSK484 |
| GSK106 |
| Physical and chemical properties for GSK484 | |
| Molecular weight | 473.2 |
| Molecular formula | C27H31N5O3 |
| IUPAC name | (3-amino-4-hydroxy-piperidin-1-yl)-(8-(7-(cyclopropyl-methyl)-7-aza-bicyclo[4.3.0]nona-1(6),2,4,8-tetraen-8-yl)-5-methoxy-7-methyl-7,9-diaza-bicyclo[4.3.0]nona-1,3,5,8-tetraen-3-yl)-methanone |
| MollogP | 2.665 |
| PSA | 71.66 |
| No. of chiral centres | 2 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 3 |
| Physical and chemical properties for GSK106 (Negative Control) | |
| Molecular weight | 401,51 |
| Molecular formula | C24H27N5O |
| IUPAC name | (3-aminopiperidin-1-yl)(2-(1-ethyl-1H-indol-2-yl)-1-methyl-1H-benzo[d]imidazol-6-yl)methanone |
| MollogP | 3.13 |
| PSA | 47.78 |
| No. of chiral centres | 1 |
| No. of rotatable bonds | 4 |
| No. of hydrogen bond acceptors | 4 |
| No. of hydrogen bond donors | 3 |
SMILES:
GSK484: CN1C2=C(OC)C=C(C(N3C[C@@H]([C@@H](CC3)O)N)=O)C=C2N=C1C4=CC5=C(C=CC=C5)N4CC6CC6
GSK106: CCN1C(C2=NC3=CC=C(C(N4CCCC(N)C4)=O)C=C3N2C)=CC5=C1C=CC=C5
InChI:
GSK484: InChI=1S/C27H31N5O3/c1-30-25-20(11-18(13-24(25)35-2)27(34)31-10-9-23(33)19(28)15-31)29-26(30)22-12-17-5-3-4-6-21(17)32(22)14-16-7-8-16/h3-6,11-13,16,19,23,33H,7-10,14-15,28H2,1-2H3/t19-,23+/m0/s1
GSK106: InChI=1S/C24H27N5O/c1-3-29-20-9-5-4-7-16(20)13-22(29)23-26-19-11-10-17(14-21(19)27(23)2)24(30)28-12-6-8-18(25)15-28/h4-5,7,9-11,13-14,18H,3,6,8,12,15,25H2,1-2H3
InChIKey:
GSK484: BDYDINKSILYBOL-WMZHIEFXSA-N
GSK106: ZDERDCQPANYYQW-UHFFFAOYSA-N
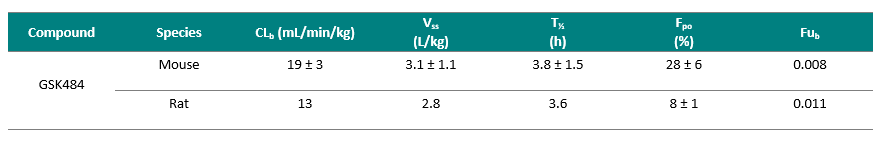
GSK Data - shared with SGC February y2016.
All animal studies were ethically reviewed and carried out in accordance with Animals (Scientific Procedures) Act 1986 and the GSK Policy on the Care, Welfare and Treatment of Animals.