This probe is available from Sigma and Cayman Chemical.
This probe (as hydrochloride) is available from Cayman Chemical.
| Probe |
 |
I-CBP112 |
CREBBP (CBP) and EP300 are general transcriptional co-activators, which are involved in many biological processes like maintenance of genomic stability by affecting DNA replication and DNA repair as well as cell growth, transformation and development. They also plays and essential role in neuronal plasticity/ memory formation, hematopoiesis and energy homeostasis as demonstrated in a variety of mouse models. By acetylation of non-histone proteins CREBBP can have a positive or negative effect on transcriptional regulation affecting protein- protein interactions, protein-DNA interactions, nuclear retention or protein half-life.
Mutations of CREBBP and EP300 lead to Rubinstein-Taybi syndrome (RTS), characterised by growth retardation, facial abnormalities, organ abnormalities, mental retardation and proneness to tumors. Chromosomal translocations of CREBBP or EP300 with MOZ, MLL have been observed in acute myeloid leukemia. CREBBP has also been associated with Amyotrophic lateral sclerosis (ALS), Lou Gherig’s disease, a neurodegenerative disease with progressive degeneration of motor neurons in the brain and spinal cord, Alzheimer's disease and polyglutamine diseases such as Spinal and Bulbar Muscular Atrophy and Huntington’s disease.
We have developed an inhibitor, I-CBP112, against the CREBBP and EP300 Bromodomains.
| In vitro Potency | |
|---|---|
| Assay | IC50/Kd |
| CREBBP (Alphascreen) | 0.170 |
| CREBBP (BLI) | 0.170 |
| CREBBP (ITC) | 0.151 |
| EP300 (ITC) | 0.625 |
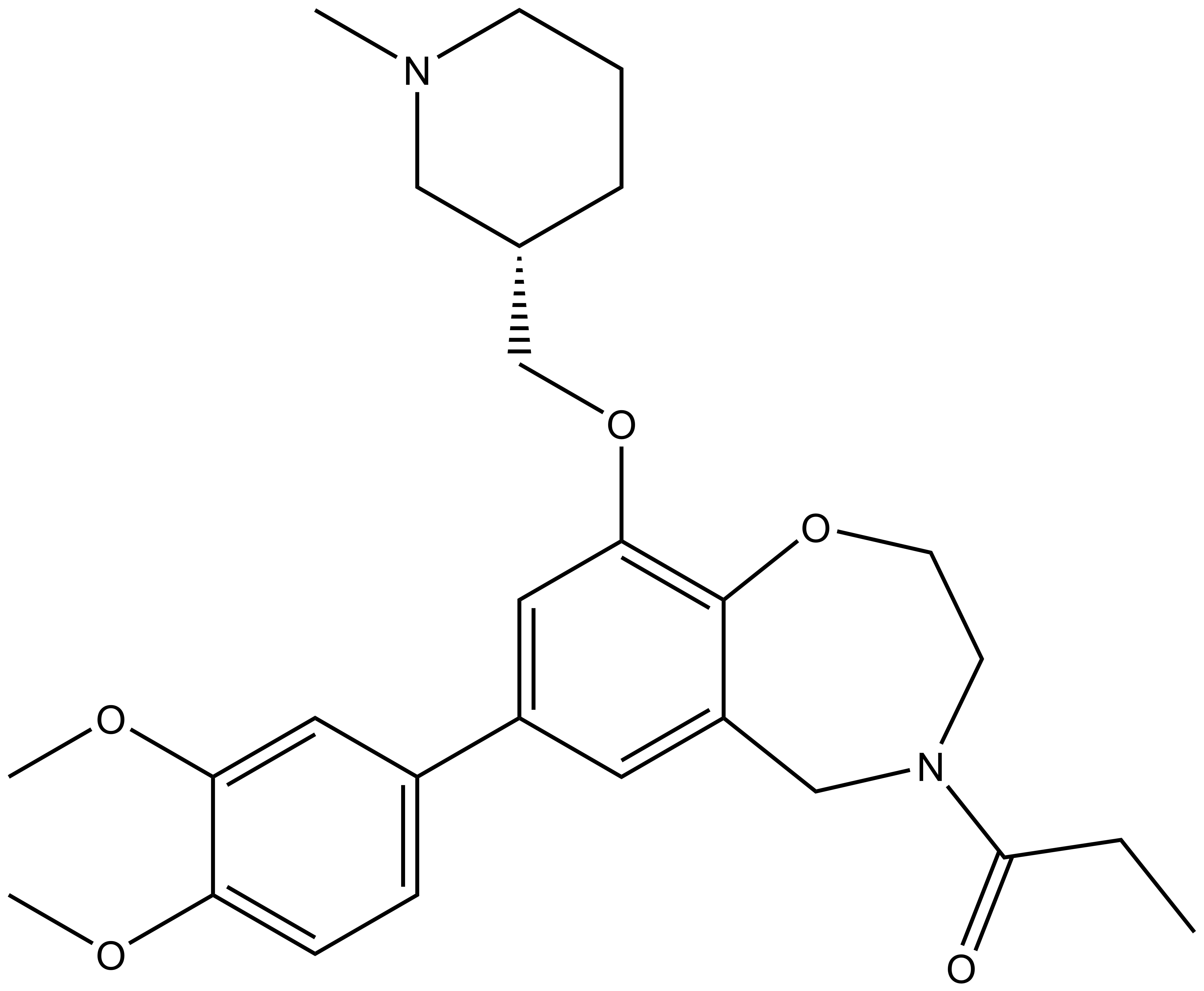
| I-CBP112 |
 |
| 1-[7-(3,4-dimethoxyphenyl)-9-{[(3S)-1-methylpiperidin-3-yl]methoxy}-2,3,4,5-tetrahydro-1,4-benzoxazepin-4-yl]propan-1-one Click here to download SDF file |
| Physical and chemical properties | |
|---|---|
| Molecular weight | 468.3 |
| clogP | 4.5 |
| PSA | 50.2 |
| Storage | 2-8°C as powder. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | Soluble in DMSO at least up to 50 mM |
Selectivity against other Bromodomains
BLI@ 0.2 and 1 µM:
No interaction was detected by BLI for the selectivity panel comprising bromodomains:
ATAD2
BAZ2B
BRD2(2)
BRD4 (1)
PB1(5)
PCAF
PHIP(2)
TIF1α
Against the entire bromodomain family, weak cross-reactivity was only observed for the BET bromodomains.
| Probe |
 |
I-CBP112 |
CREBBP (3x bromodomain) (FRAP assay) - Accelerated FRAP recovery at 1 µM.
No significant cytotoxicity up to 50 μM in U2OS cells.
Generation of a selective small molecule inhibitor of the CBP/p300 bromodomain for leukemia therapy.
Picaud S, Fedorov O, Thanasopoulou A, Leonards K, Jones K, Meier J, Olzscha H, Monteiro O, Martin S, Philpott M, Tumber A, Filippakopoulos P, Yapp C, Wells C, Hing Che K, Bannister A, Robson S, Kumar U, Parr N, Lee K, Lugo D, Jeffrey P, Taylor S, Vecellio ML, Bountra C, Brennan P, O'Mahony A, Velichko S, Muller S, Hay D, Daniels DL, Urh M, La Thangue NB, Kouzarides T, Prinjha R, Schwaller J, Knapp S.
Cancer Res. 2015;75(23):5106-19.
I-CBP112 has been co-crystallised with CREBBP and is deposited in the PDB - 4NR6