The probe PFI-6 is available from Sigma and Cayman Chemicals.
The negative control PFI-6N is available from Sigma.
| Probe | Negative control | |
 |
|  |
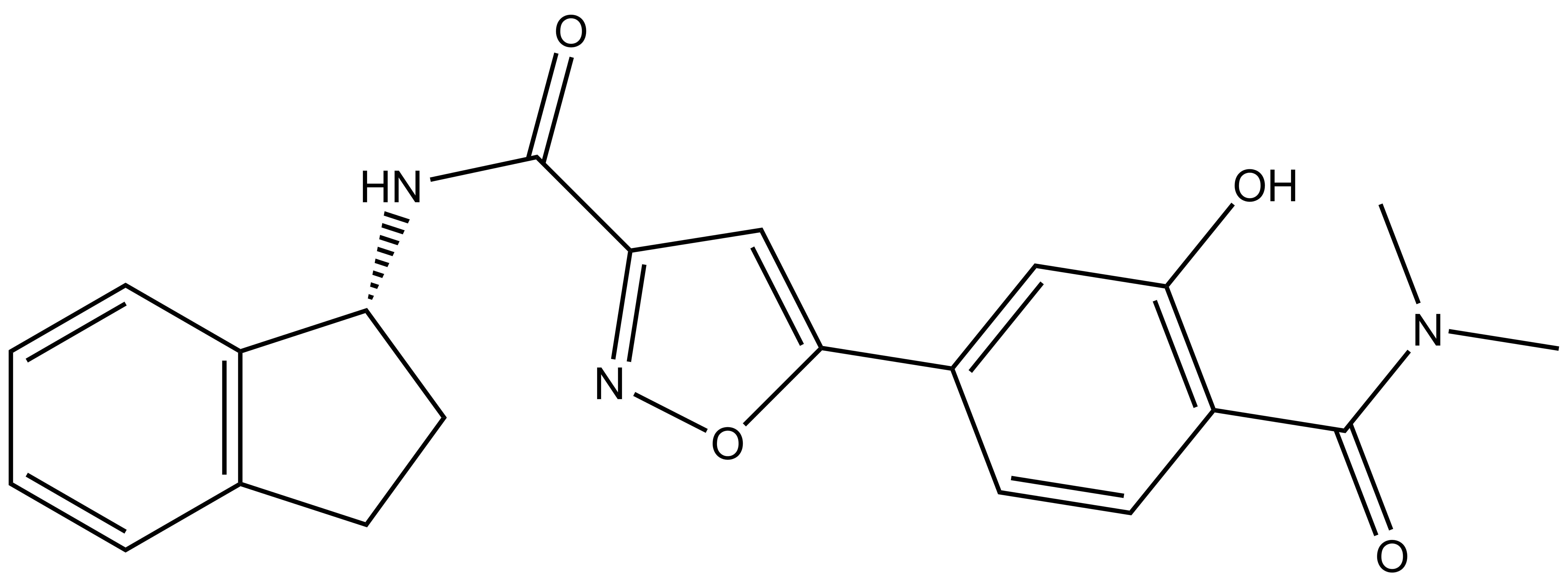
PFI-6 |
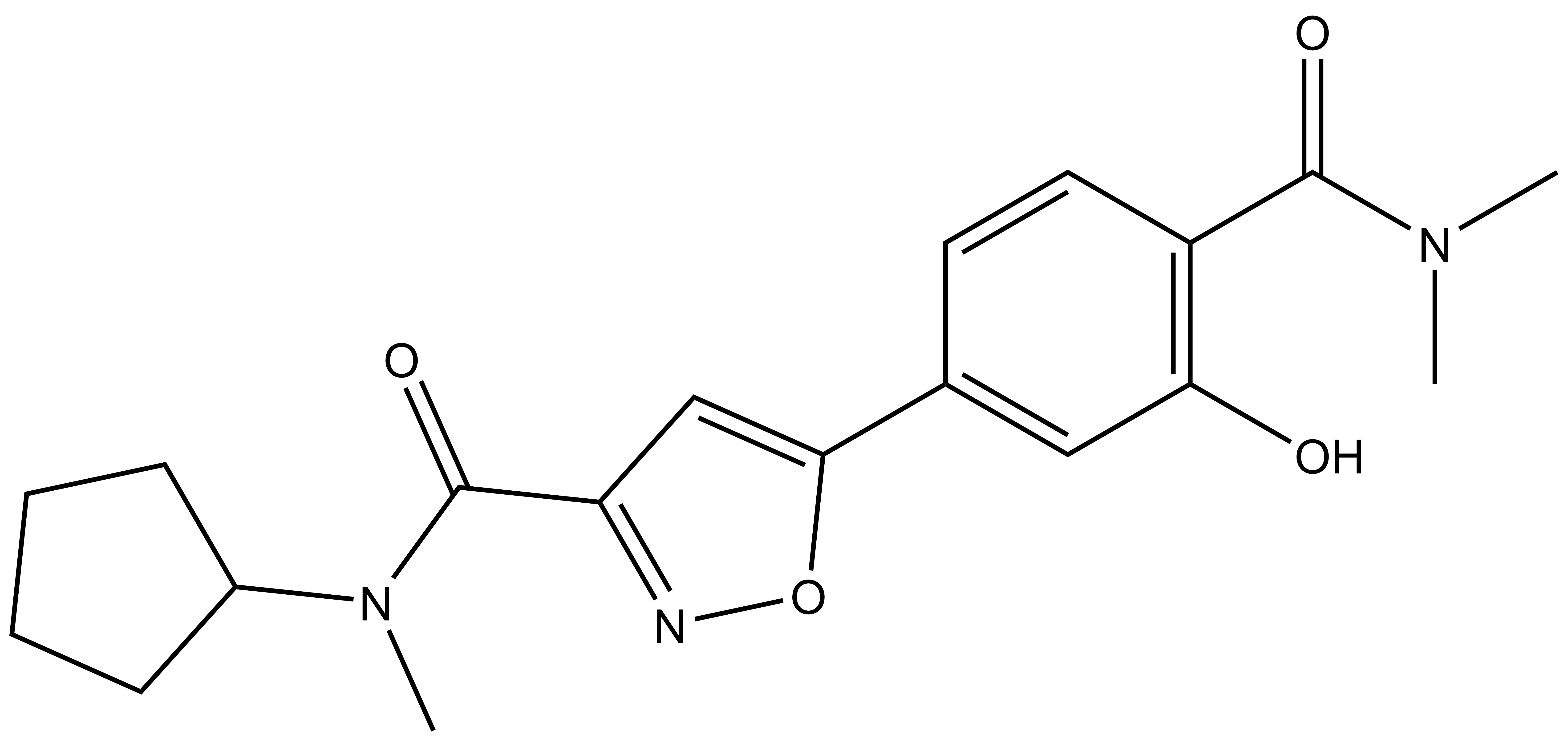
| PFI-6N |
Increasing numbers of papers describing YEATS biology are emerging, and there is a growing need for high quality chemical probes that can be used to study YEATS domains. A collaboration between Pfizer and the SGC has resulted in the discovery of PFI-6, a potent inhibitor of MLLT1/3. PFI-6 has a unique chemotype relative to the current MLLT1/3 chemical probe NVS-MLLT-1. PFI-6N has also been developed as a structurally similar negative control compound.
| Probe |
 |
PFI-6 |
| Physical and chemical properties for PFI-6 | |
| Molecular weight | 391.43 |
| Molecular formula | C22H21N3O4 |
| IUPAC name | (R0-N-(2,3-dihydro-1H-inden-1-yl)-5-(4-(dimethylcarbamoyl)-3-hydroxyphenyl)isoxazole-3-carboxamide |
| MollogP | 2.6 |
| PSA | 91.23 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 7 |
| No. of hydrogen bond donors | 2 |
| Storage | store as a dry poweder or as DMSO stock solutions (10mM) at -20 °C |
| Dissolution | Soluble in DMSO up to 10mM |
| Negative control |
 |
PFI-6N |
| Physical and chemical properties for PFI-6N | |
| Molecular weight | 357.41 |
| Molecular formula | C19H23N3O4 |
| IUPAC name | N-cyclopentyl-5-(4-(dimethylcarbamoyl)-3-hydroxyphenyl)-N-methylisoxazole-3-carboxamide |
| MollogP | 1.86 |
| PSA | 82.44 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 7 |
| No. of hydrogen bond donors | 1 |
| Storage | Store as a dry powder or as DMSO stock solutions (10mM) at -20 °C |
| Dissolution | Soluble in DMSO up to 10mM |
SMILES:
PFI-6: O=C(C1=C(C=C(C=C1)C2=CC(C(N[C@@H]3CCC4=C3C=CC=C4)=O)=NO2)O)N(C)C
PFI-6N: O=C(C1=C(C=C(C=C1)C2=CC(C(N(C3CCCC3)C)=O)=NO2)O)N(C)C
InChI:
PFI-6: 1S/C22H21N3O4/c1-25(2)22(28)16-9-7-14(11-19(16)26)20-12-18(24-29-20)21(27)23-17-10-8-13-5-3-4-6-15(13)17/h3-7,9,11-12,17,26H,8,10H2,1-2H3,(H,23,27)/t17-/m1/s1
PFI-6-N: 1S/C19H23N3O4/c1-21(2)18(24)14-9-8-12(10-16(14)23)17-11-15(20-26-17)19(25)22(3)13-6-4-5-7-13/h8-11,13,23H,4-7H2,1-3H3
InChIKey:
PFI-6: IXWUILRSNIQHDM-QGZVFWFLSA-N
PFI-6-N:CKEICVFLYGXFOP-UHFFFAOYSA-N
PFI-6 shows potent activity on MLLT1/3 (Table 1). PFI-6 is universally inactive in the SGC Bromodomain panel, and showed no activity in an Invitrogen panel of 40 kinases (screening conducted at 10µM). Additionally, PFI-6 showed no activity in a panel of 25 PDEs, ion channels and GPCRs >50µM. The negative control, PFI-6N, was not reactive against MLLT1 >30µM, MLLT3 >30µM, YEATS2 >30µM and YEATS4 >30µM.
| MLLT1 | MLLT3 | YEATS2 | YEATS4 | |
| HTRF IC50 (µM) | 0.14 | 0.16 | >40 | >40 |
| BLI Kd (µM) | 0.11 | 0.11 | n.d. | n.d. |
| ITCKD (µM) | 0.082 | 0.076 | n.d. | n.d. |
| Tm Shift (°C) | 3.61 | 5.13 | 0.00 | -0.02 |
Table 1: Potency Against Target Family
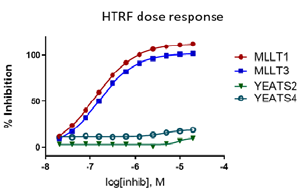
Figure 1: PFI-6 specfically inhibits MLLT1 and MLLT3.
In the NanoBRET cellular target engagement assay, PFI-6 displayed potent inhibition, with an average IC50 value of 0.76 μM (±0.1) (Figure 1). In comparison, PFI-6N had no inhibitory properties (up to 30 μM) (Figure 1). Further in cell validation using a Fluorescence Recovery After Photobleaching (FRAP) assay was used to confirm target inhibition by PFI-6 (Figure 2) .
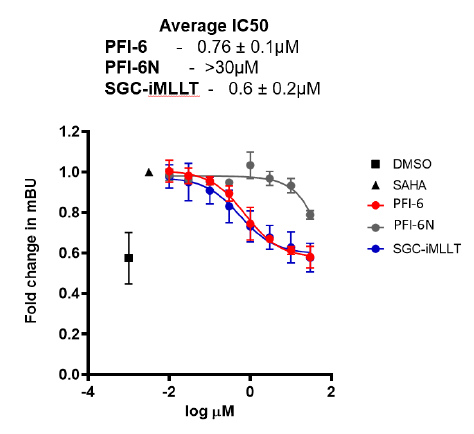
Figure 1: A NanoBRET assay was used to determine MLLT3 target engagement in cells.
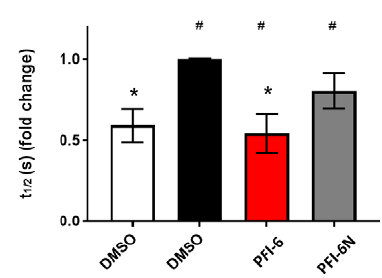
Figure 2: Confirmation of MLLT1 target engagement in cells by Fluorescence Recovery After Photobleaching analysis.
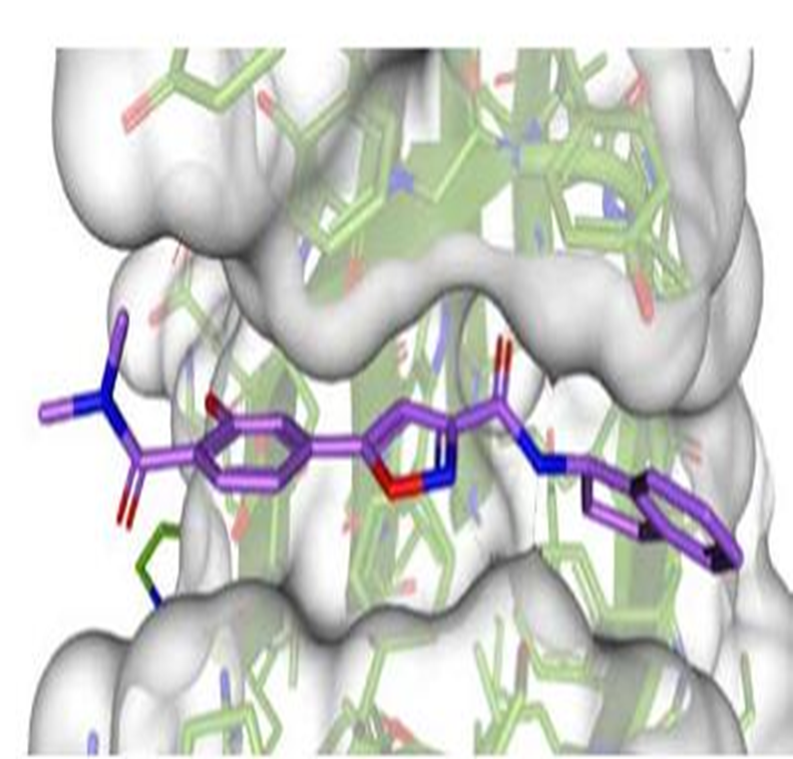
The figure below shows PFI-6 is soaked into the YEATS domain. PDB codes will be added when they become available.