PFI-7
Chemical probe for GID4, substrate-recognition subunit of the CTLH E3 ubiquitin-protein ligase complex
The probe PFI-7 (hydrochloride) is available at Sigma and Tocris.
The inactive control PFI-7N (hydrochloride) is available at Sigma.
-
overview
-
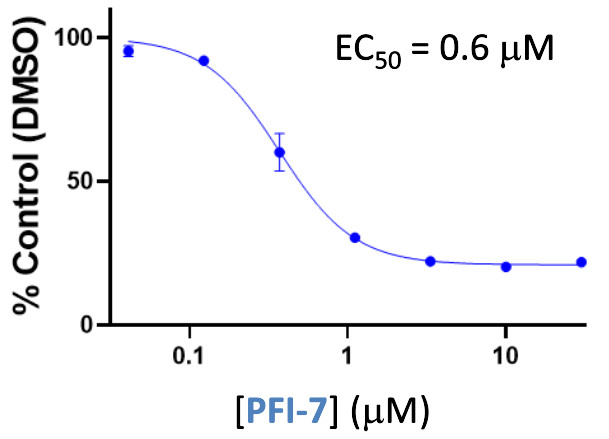
in vitro potency
-
cell based assay data
-
references
-
co-crystal structures
overview
| Probe | | Negative control |
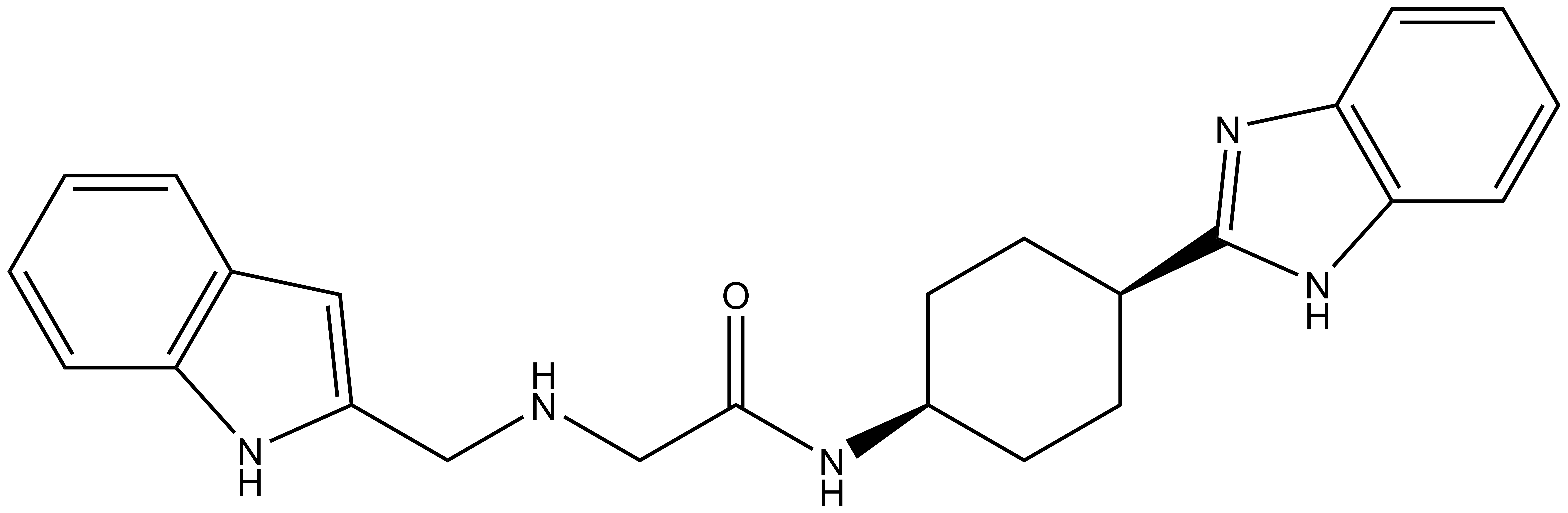 | | 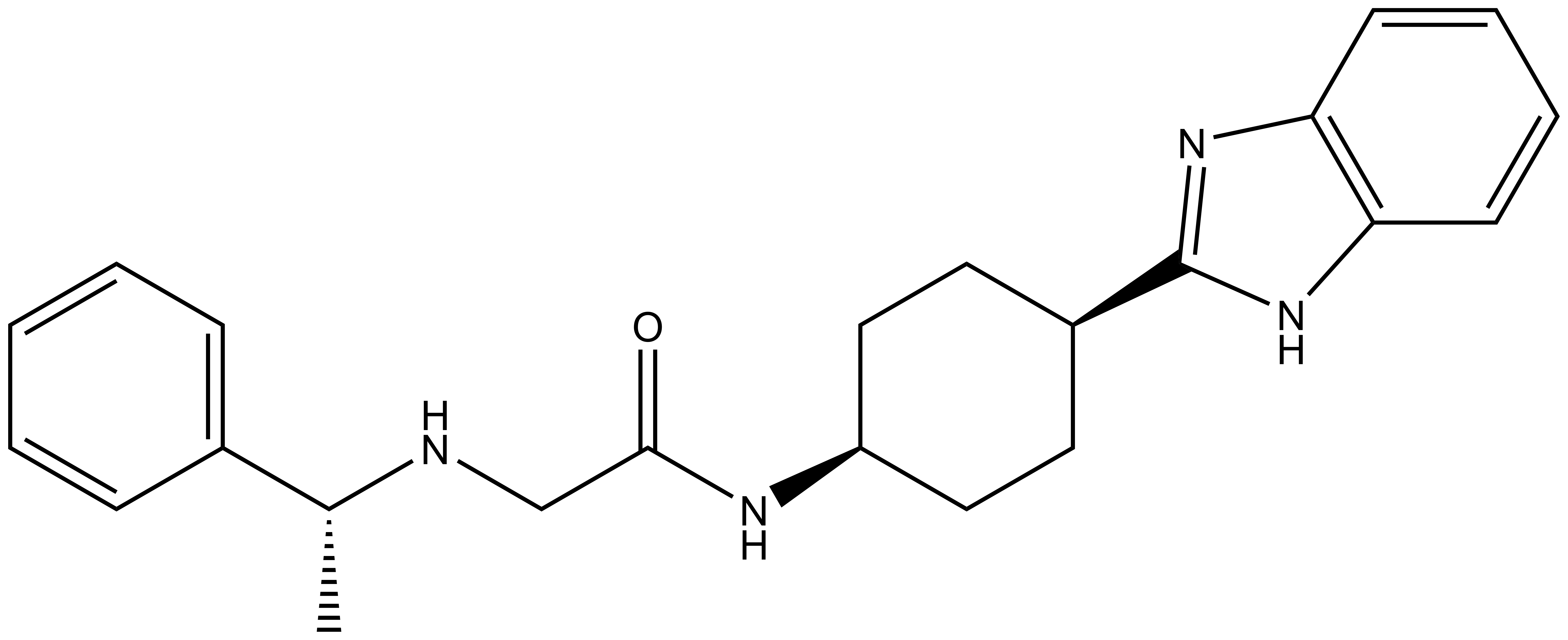 |
PFI-7 | | PFI-7N |
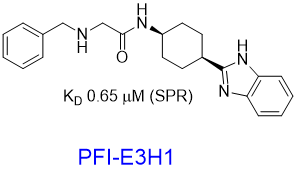
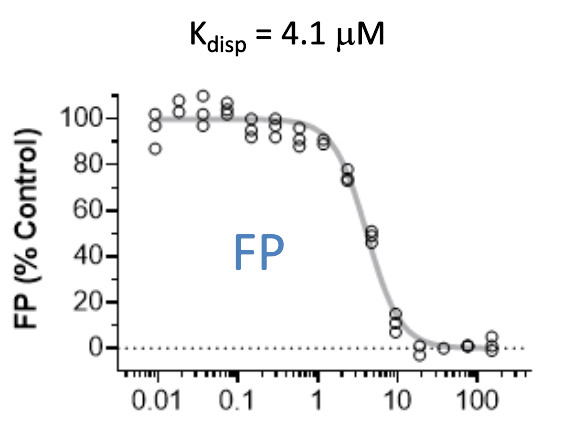
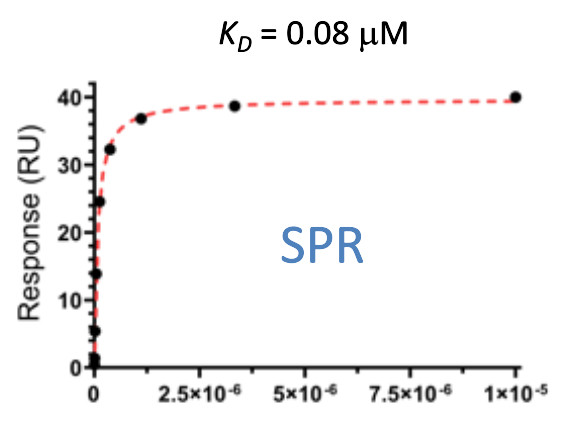
Pfizer in collaboration with the SGC have developed PFI-7, a potent, cell active chemical probe for the E3 ligase GID4. PFI-7 binds potently to GID4 with KD = 0.08 μM (SPR) and displaces the known degron1 peptide in a NanoBRETTM assay with EC50 = 0.6 μM. PFI-7N is a closely related negative control with KD = 5 μM (SPR). A co-crystal structure has been deposited.
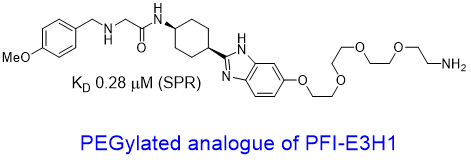
We have further developed a handle PFI-E3H1 and a PEGylated analogue to show that the handle tolerates a substitution. These findings offers opportunities to synthesize proximity-inducing or degrader modalities2.
cell based assay data
A NanoBRET assay was used to show target engagement in cells.
The interaction was between NanoLuc® tagged degrons and full-length GID4.
references
- Cheng Dong, Heng Zhang, Li Li, Wolfram Tempel, Peter Loppnau & Jinrong Min. Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway. Nature Chemical Biology 14, 466-473 (2018).
- Aleša Bricelj, Christian Steinebach, Robert Kuchta, Michael Gütschow, and Izidor Sosič. E3 Ligase Ligands in Successful PROTACs: An Overview of Syntheses and Linker Attachment Points, https://doi.org/10.3389/fchem.2021.707317 ; Milka Kostic. Targeted Protein Degradation and Proximity-Based Pharmacology, https://doi.org/10.5281/zenodo.5534371 .
- Dominic D.G. Owens et al., A chemical probe to modulate human GID4 Pro/N-degron interactions. https://pubmed.ncbi.nlm.nih.gov/38773330/
- Aliakbar K Yazdi et al., Chemical tools for the Gid4 subunit of the human E3 ligase C-terminal to LisH (CTLH) degradation complex. https://pubmed.ncbi.nlm.nih.gov/38516600/
co-crystal structures
Main features
- PFI-7 bound to GID4 substrate-binding pocket
- Structure overview
- Overlap with substrate peptide