This probe is available from Sigma.
| Probe | Negative control | |
 |
|  |
SGC-GSK3-1 |
| SGC-CDKL5/GSK3-1N |
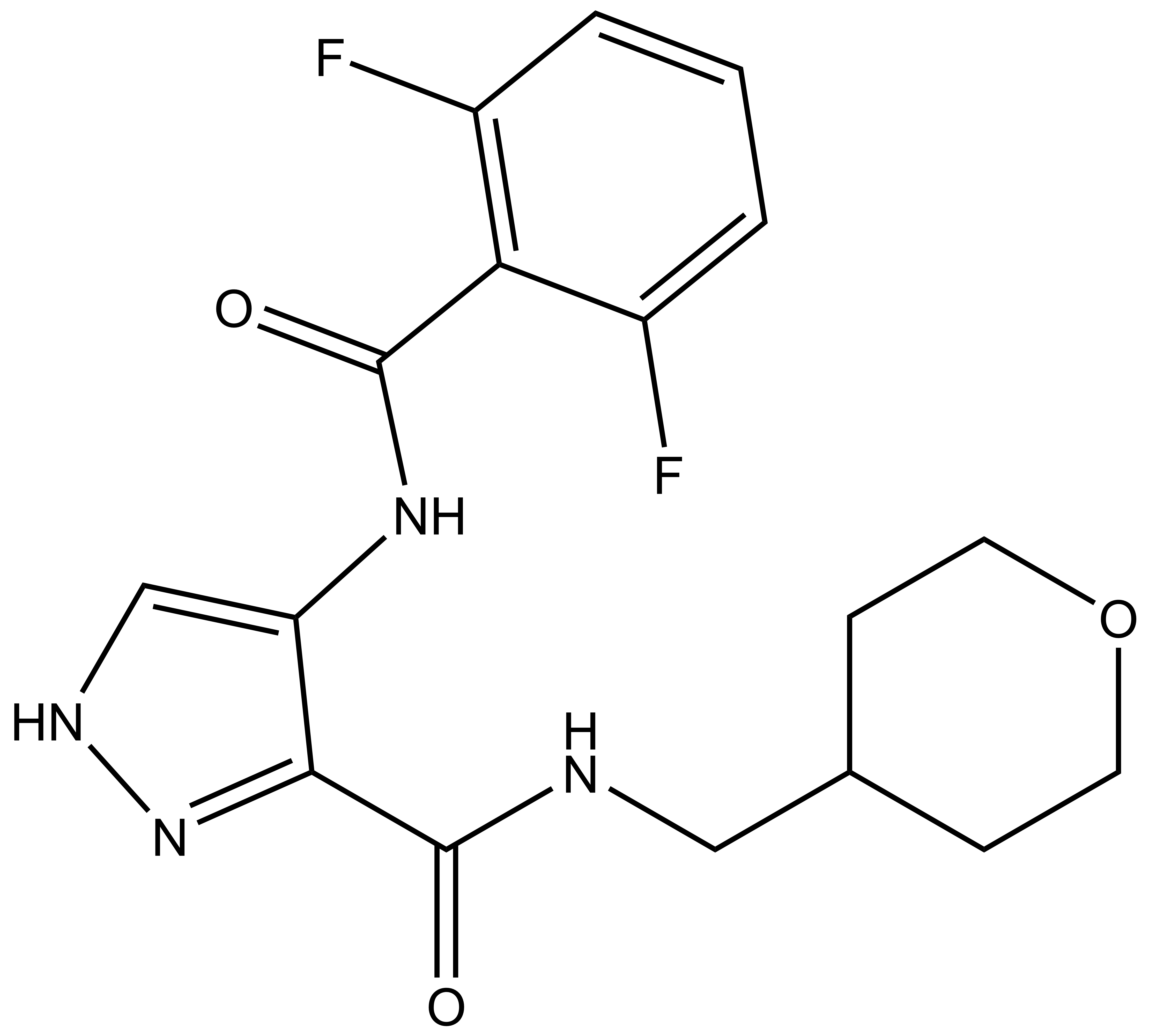
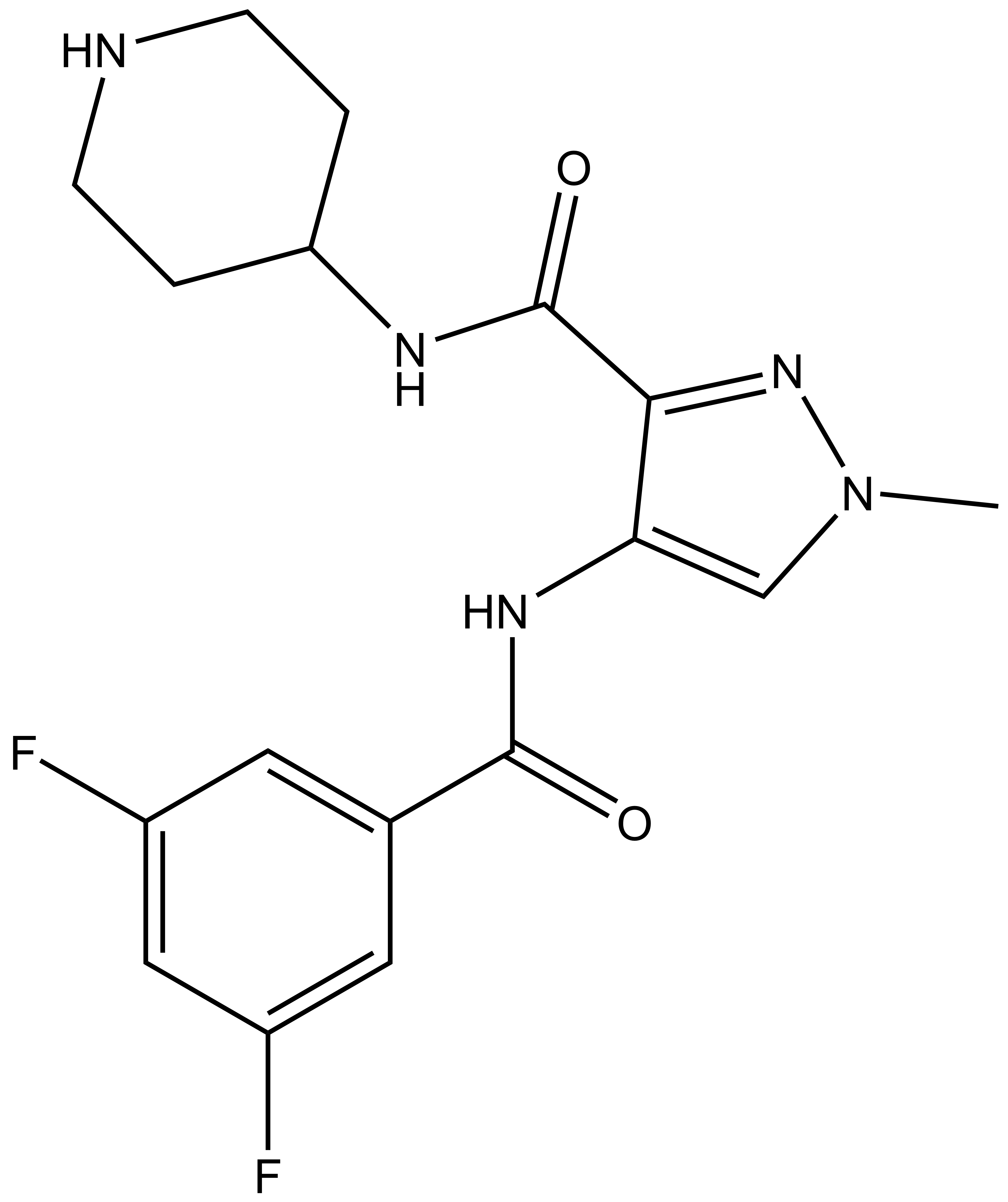
From a library of AT-7519 analogs, we identified a potent and cell-active chemical probe (SGC-GSK3-1) that inhibits glycogen synthase kinase-3 (GSK3⍺ and β). Comprehensive evaluation of kinome-wide selectivity confirmed that this GSK3 probe demonstrates remarkable selectivity. A structurally similar analog (SGC-CDKL5/GSK3-1N) was characterized as a negative control that does not bind to GSK3⍺ or GSK3β in corresponding cellular target engagement assays. At nanomolar concentrations, our GSK3 chemical probe promoted motor neuron survival when iPSC-derived motor neurons were exposed to ER stress. When used at an appropriate concentration (<500 nM) in cells, SGC-GSK3-1 is exquisitely selective for GSK3⍺ or GSK3β. Our chemical probe set can be used alongside other potent and selective, structurally divergent GSK3 inhibitors to characterize the impacts of GSK3 inhibition on downstream biology.
| Probe | Negative control | |
 |
|  |
SGC-GSK3-1 |
| SGC-CDKL5/GSK3-1N |
| Physical and chemical properties for SGC-GSK3-1 | |
| Molecular weight | 364.35 |
| Molecular formula | C17H18F2N4O3 |
| IUPAC name | 4-(2,6-difluorobenzamido)-N-((tetrahydro-2H-pyran-4-yl)methyl)-1H-pyrazole-3-carboxamide |
| ClogP | -0.52 |
| PSA | 96.11 |
| No. of chiral centers | 0 |
| No. of rotatable bonds | 7 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 3 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10 mM) are stable at -20oC |
| Dissolution | Soluble in DMSO up to 10 mM |
| Physical and chemical properties for SGC-CDKL5/GSK3-1N | |
| Molecular weight | 363.37 |
| Molecular formula | C17H19FN5O2 |
| IUPAC name | 4-(3,5-difluorobenzamido)-1-methyl-N-(piperidin-4-yl)-1H-pyrazole-3-carboxamide |
| ClogP | -0.32 |
| PSA | 88.05 |
| No. of chiral centers | 0 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 3 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10 mM) are stable at -20oC |
| Dissolution | Soluble in DMSO up to 10 mM |
SMILES:
SGC-GSK3-1: O=C(NCC1CCOCC1)C2=NNC=C2NC(C3=C(C=CC=C3F)F)=O
SGC-CDKL5/GSK3-1N: O=C(NC1CCNCC1)C2=NN(C=C2NC(C3=CC(F)=CC(F)=C3)=O)C
InChI:
SGC-GSK3-1: InChI=1S/C17H18F2N4O3/c18-11-2-1-3-12(19)14(11)16(24)22-13-9-21-23-15(13)17(25)20-8-10-4-6-26-7-5-10/h1-3,9-10H,4-8H2,(H,20,25)(H,21,23)(H,22,24)
SGC-CDKL5/GSK3-1N: InChI=1S/C17H19F2N5O2/c1-24-9-14(22-16(25)10-6-11(18)8-12(19)7-10)15(23-24)17(26)21-13-2-4-20-5-3-13/h6-9,13,20H,2-5H2,1H3,(H,21,26)(H,22,25)
InChIKey:
SGC-GSK3-1: OCQCKUTZHBQNEI-UHFFFAOYSA-N
SGC-CDKL5/GSK3-1N: ODZBDODDFLVDQZ-UHFFFAOYSA-N
SGC-GSK3-1 was profiled in the DiscoverX scanMAX assay against 403 wild-type kinases at 1 μM. Only 5 kinases showed PoC <10 giving an S10(1 μM) = 0.012. When the PoC <35 fraction was examined, 15 kinases were included (S35(1 μM) = 0.037). Potential off-targets within the S35(1 μM) fraction were tested via biochemical enzymatic/binding assays plus NanoBRET target engagement assays for CDKL5, GSK3⍺, GSK3β, and DYRK1B. Data corresponding with off-target kinase activity is shown in the table below.
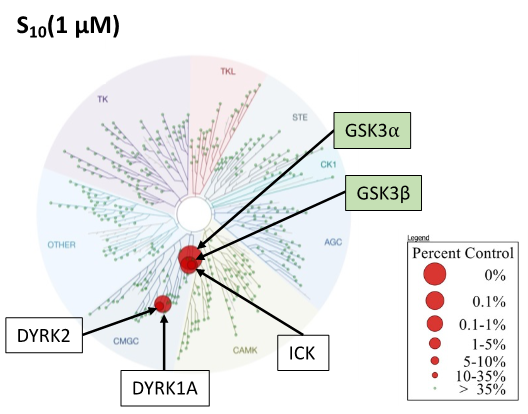
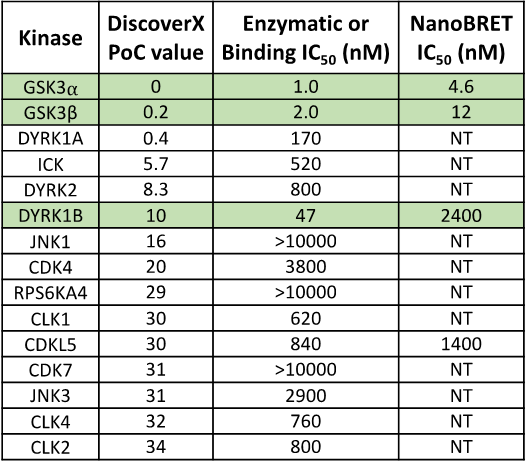
Figure 2: SGC-GSK3-1 was profiled in the DiscoverX scanMAX assay against 403 wild-type kinases at 1 μM and off-target kinases inhibited PoC <35 were tested in an orthogonal assay. Rows colored green are GSK3⍺, GSK3β, and DYRK1B. No other kinases demonstrate enzymatic IC50 values within 30-fold of the GSK3β enzymatic IC50 value.
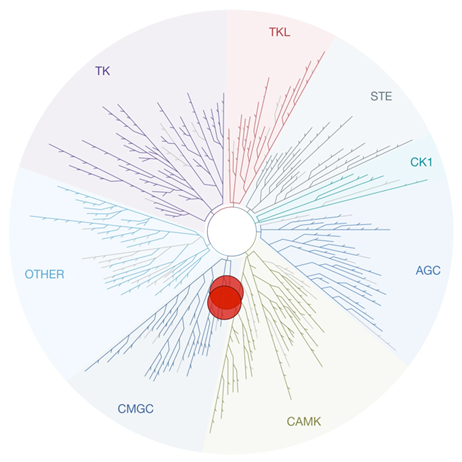
Figure 1: Kinome tree with GSK3⍺ and GSK3β highlighted as red circles. Illustration is reproduced courtesy of Eurofins DiscoverX (http://treespot.discoverx.com).
Biological activity summary:
A NanoBRET assay was utilized to assess the binding affinity of SGC-GSK3-1 to CDKL5, GSK3⍺, GSK3β, and DYRK1B. The negative control shows no binding affinity for GSK3⍺ or GSK3β.
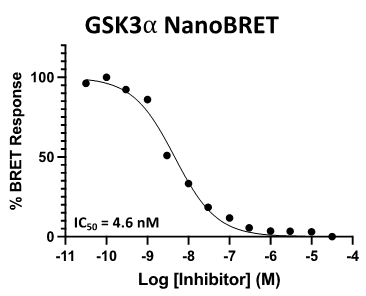
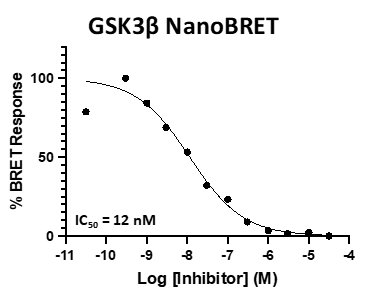
Figure 3: SGC-GSK3-1 was profiled in the GSK3⍺ and GSK3β NanoBRET assays.
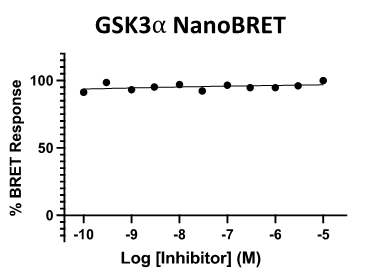

Figure 4: SGC-CDKL5/GSK3-1N was profiled in the GSK3⍺ and GSK3β NanoBRET assays.
References
Ong, H. W.; Liang, Y.; Richardson, W.; Lowry, E. R.; Wells, C. I.; Chen, X.; Silvestre, M.; Dempster, K.; Silvaroli, J. A.; Smith, J. L.; Wichterle, H.; Pabla, N. S.; Ultanir, S. K.; Bullock, A. N.; Drewry, D. H.*; Axtman, A. D.* Discovery of a potent and selective CDKL5/GSK3 chemical probe that is neuroprotective. ACS Chem Neurosci 2023, 14, 1672–1685; 10.1021/acschemneuro.3c00135.
Ong, H. W.; Liang, Y.; Richardson, W.; Lowry, E. R.; Wells, C. I.; Chen, X.; Silvestre, M.; Dempster, K.; Silvaroli, J. A.; Smith, J. L.; Wichterle, H.; Pabla, N. S.; Ultanir, S. K.; Bullock, A. N.; Drewry, D. H.*; Axtman, A. D.* A potent and selective CDKL5/GSK3 chemical probe is neuroprotective. BioRxiv 2023, doi: 10.1101/2023.02.09.527935.