| Probe | Negative control | |
 |
|  |
SGC-SMARCA-BRDVIII |
| SGC-BRDVIII-NC |
The SWI/SNF (switch/sucrose non-fermenting) chromatin remodelling complexes, BAF (BRG1/BRM-associated factor) and PBAF (polybromo-associated BAF), are crucial epigenetic regulators to control DNA accessibility, and thus largely contribute to cell proliferation and differentiation mechanisms [1]. The complexes incorporate either the related subunit SMARCA2 or SMARCA4 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 2/4), which both consist of a mutually exclusive catalytic ATPase domain and a bromodomain that reads acetylated histones on the chromatin. A unique feature of PBAF is the subunit PBRM1 (polybromo-1) containing six tandem-acting bromodomains. SWI/SNF complexes are known to be strong tumor suppressors and their dysfunctions trigger substantial oncogenic programs or deregulate cell lineage differentiation mechanisms [1].
Here, we present a new chemical probe for the SMARCA2/4 and PB1(5) bromodomains of BAF/PBAF, SGC-SMARCA-BRDVIII, that was initially designed by Genentech/Constellation [2] and then further advanced into the chemical probe platform by the SGC Frankfurt [3]. This scaffold represents the second chemical probe for the SMARCA2/4 and PB1(5) bromodomains that is based on a different chemotype than our first generation bromodomain inhibitor PFI-3 [4,5]. Moreover, this inhibitor has also recently been used to develop the PROTAC (proteolysis targeting chimera) ACBI-1 [6].
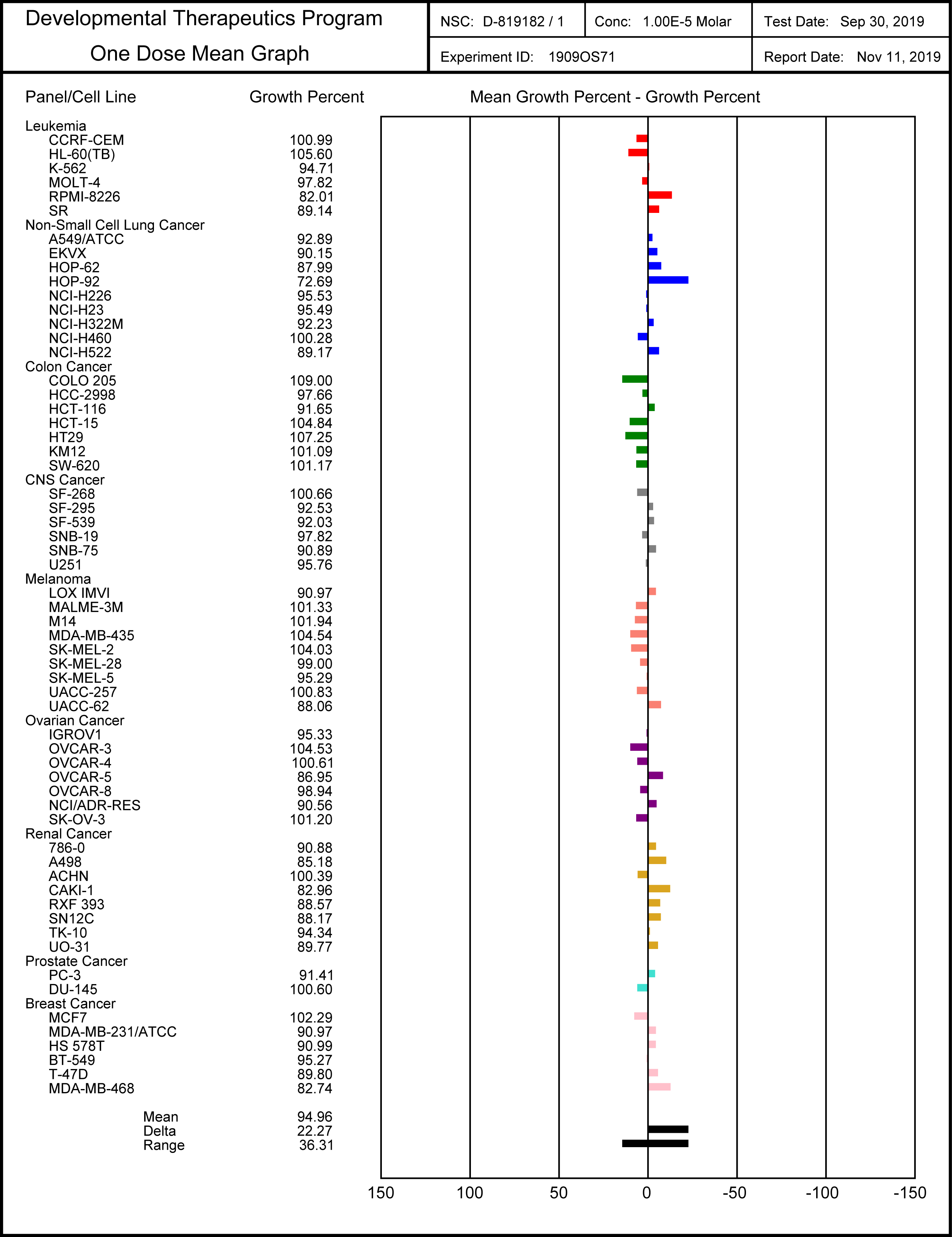
SGC-SMARCA-BRDVIII binds potently to the SMARCA2/4 and PB1(5) bromodomains with a KD(ITC) of 35, 36 and 13 nM. It is selective within the other bromodomain families and shows no off-targets activity on 85 protein kinases screened using temperature shift binding assays. This compound is non-toxic as demonstrated by the NCI-60 human tumor cell lines screen, but it shows remarkably cellular activity in an adipogenesis cell differentiation assay with an EC50 of < 1.0 µM.
Of special note is that SGC-SMARCA-BRDVIII outperformed the chemical probe PFI-3 in that particular assay, and therefore, we advise to use SGC-SMARCA-BRDVIII and PFI-3 to confirm the results, when elucidating the biological role of the SWI/SNF bromodomains.
In addition, a negative control compound, SGC-BRDVIII-NC, is provided, which showed no cellular activity. If interested, a chemogenomic tool compound, SGC-pan-BRDVIII, that additionally hits the PB1(2,3) members with a KD(ITC) of 200-400 nM can be inquired [3].
Potency Against Target Family
| Bromodomain | SGC-SMARCA-BRDVIII KD (nM) |
| SMARCA2 | 35 |
| SMARCA4 | 36 |
| PB1(5) | 13 |
| PB1(2) | 3655 |
| PB1(3) | 1963 |
Table 1: Screening SGC-SMARCA-BRDVIII against selected targets.
Selectivity
Selectivity of the SGC-SMARCA-BRDVIII probe within the bromodomain families was confirmed by an in-house thermal shift panel containing 25 bromodomains. No activity was also observed on 85 protein kinases screened in an in-house DSF panel.
Selectivity Dosage
To minimize the chance of off-target effects, we recommend that a concentration of no higher than 10 µM should be used in cell-based assays.
Cellular Activity
The formation from 3T3-L1 mouse fibroblasts into adipocytes was impaired with an EC50 below 1.0 µM upon treatment with SGC-SMARCA-BRDVIII.
| Probe | Negative control | |
 |
|  |
SGC-SMARCA-BRDVIII |
| SGC-BRDVIII-NC |
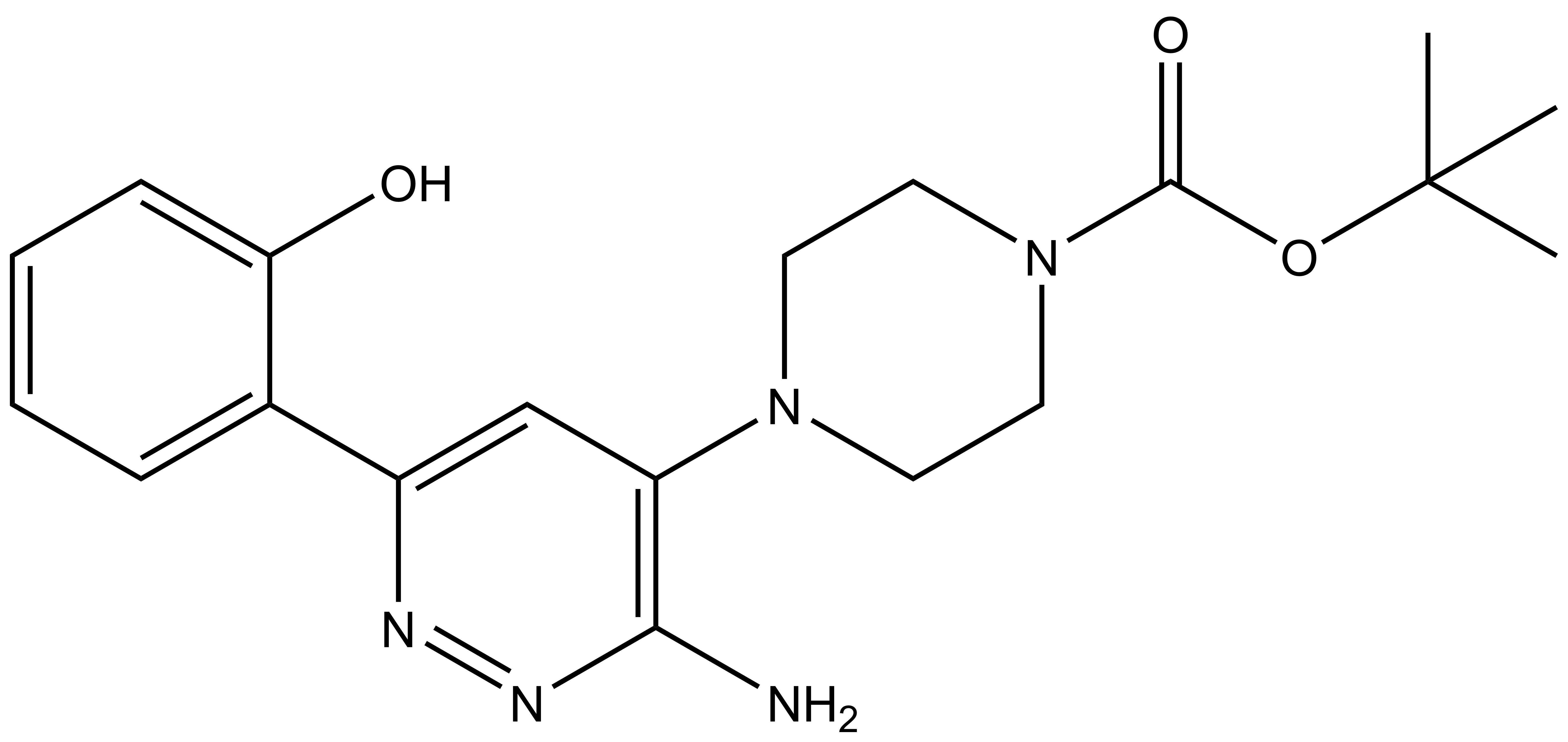
| SGC-SMARCA-BRDVIII | |
| Physical and chemical properties | |
| Molecular weight | 380.45 g/mol |
| Molecular formula | C19H25N5O3 |
| IUPAC name | tert-butyl 4-[3-amino-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazine-1-carboxylate |
| logP | 1.82 |
| TPSA | 104.8 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 5 |
| No. of hydrogen bond acceptors | 5 |
| No. of hydrogen bond donors | 2 |
| Storage | Stable as powder at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | DMSO (up to 50 mM) |
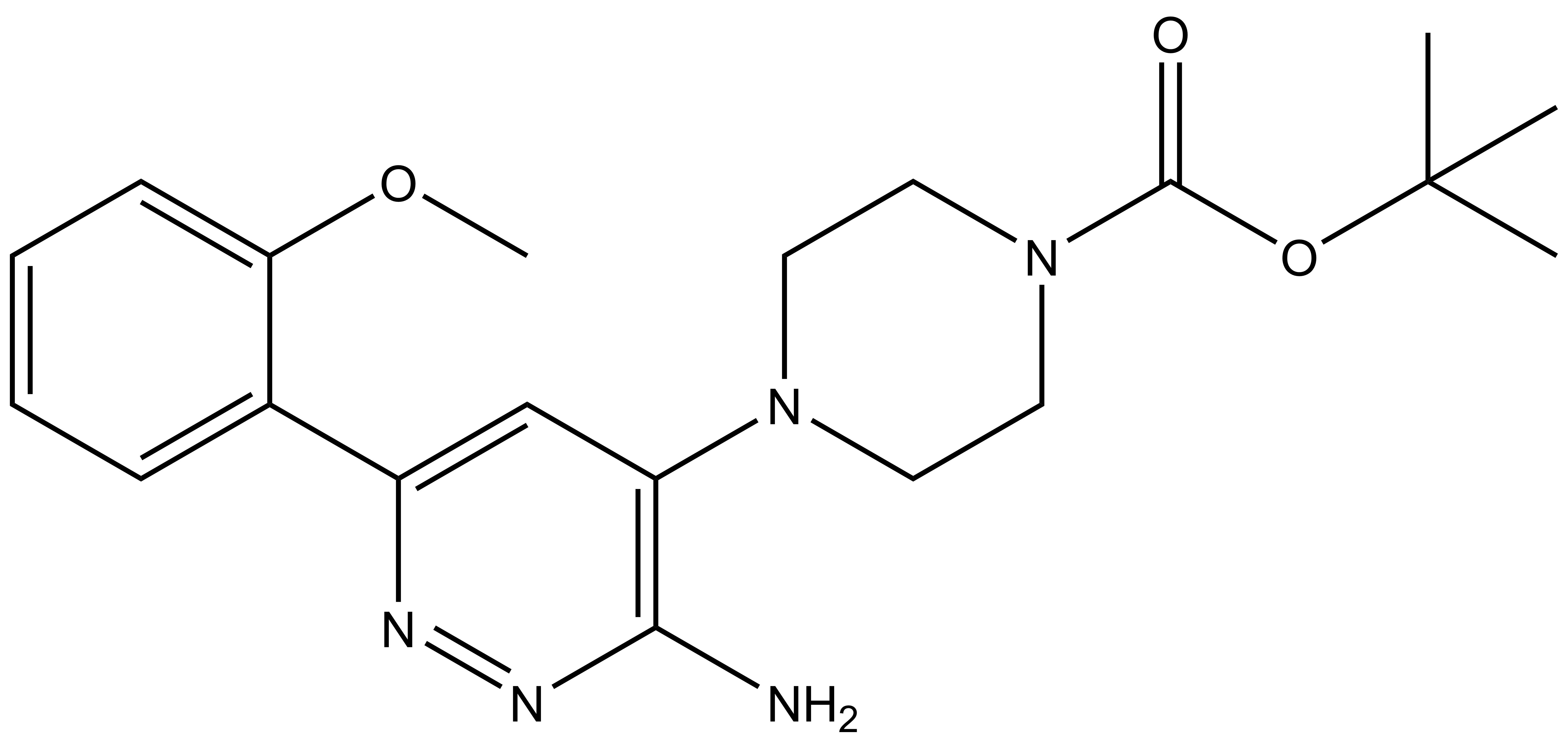
| SGC-BRDVIII-NC | |
| Physical and chemical properties | |
| Molecular weight | 385.21 g/mol |
| Molecular formula | C20H27N5O3 |
| IUPAC name | tert-butyl-4-[3-amino-6-(2-methoxyphenyl)yridazine-4-yl]piperazine-1-carboxylate |
| clogP | 2.02 |
| TPSA | 93.8 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 5 |
| No. of hydrogen bond donors | 1 |
| Storage | Stable as powder at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | DMSO (up to 50 mM) |
SMILES:
SGC-SMARCA-BRDVIII: OC1=CC=CC=C1C2=CC(N3CCN(CC3)C(OC(C)(C)C)=O)=C(N=N2)N
SGC-BRDVIII-NC: NC(N=NC(C1=CC=CC=C1OC)=C2)=C2N3CCN(CC3)C(OC(C)(C)C)=O
InChI:
SGC-SMARCA-BRDVIII: InChI=1S/C19H25N5O3/c1-19(2,3)27-18(26)24-10-8-23(9-11-24)15-12-14(21-22-17(15)20)13-6-4-5-7-16(13)25/h4-7,12,25H,8-11H2,1-3H3,(H2,20,22)
SGC-BRDVIII-NC:InChI=1S/C20H27N5O3/c1-20(2,3)28-19(26)25-11-9-24(10-12-25)16-13-15(22-23-18(16)21)14-7-5-6-8-17(14)27-4/h5-8,13H,9-12H2,1-4H3,(H2,21,23)
InChIKey:
SGC-SMARCA-BRDVIII: AQTNUGRRZDRZIA-UHFFFAOYSA-N
SGC-BRDVIII-NC: YOBVMDSDYHOOLN-UHFFFAOYSA-N
In-house DSF panel revealed no off-targets for SGC-SMARCA-BRDVIII outside the BRD subfamily VIII. The negative control compound, SGC-BRDVIII-NC is completely inactive on all tested targets.
| Bromodomains | SGC-SMARCA-BRDVIII (ΔTm°C)a,b | ||
| TAF1L(1) | -0.2± 0.1 | ATAD2 | 0.4± 0.1 |
| TAF1L(2) | -0.1± 0.1 | EP300 | 0.0± 0.1 |
| BRD2(1) | 0.0± 0.2 | BRD4(1) | 0.7± 0.1 |
| PB1(1) | 0.9± 0.1 | BRD3(2) | -0.1± 0.1 |
| PB1(2) | 2.5± 0.1 | BRPF1A | 0.2± 0.1 |
| PB1(3) | 2.7± 0.2 | BRD1 | 0.2± 0.1 |
| PB1(4) | 0.7± 0.1 | TRIM33B | 0.1± 0.1 |
| PB1 | 11.3± 0.3 | SP110A | -0.1± 0.1 |
| PB1 | 0.3± 0.1 | PCAF | 0.0± 0.1 |
| SMARCA2 | 7.7± 0.2 | WDR9 | -0.4± 0.1 |
| SMARCA4 | 7.4± 0.1 | BRDT(1_2) | 0.1± 0.1 |
| CREBBP | -0.1± 0.1 | ||
Table 1: Screening SGC-SMARCA-BRDVIII against a panel of bromodomains.
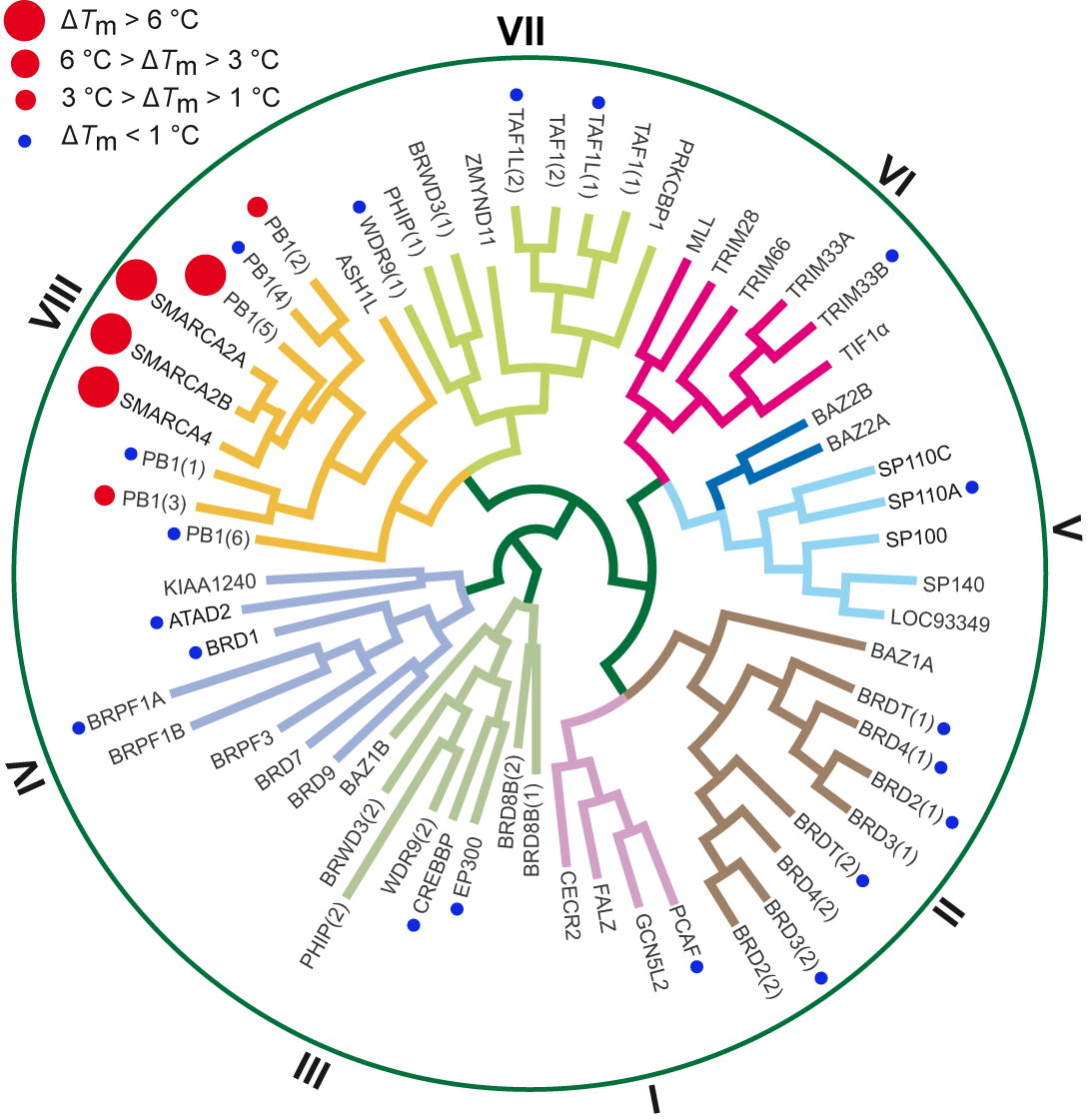
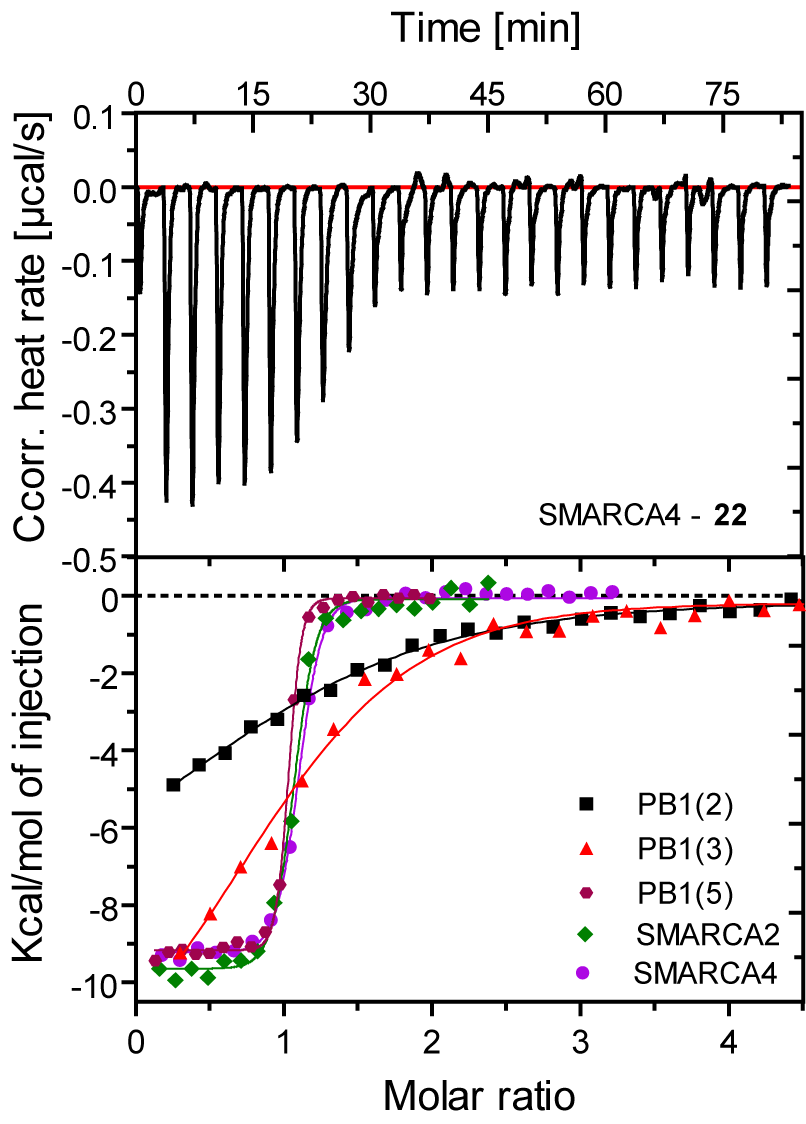
Figure 1: SGC-SMARCA-BRDVIII (22) binds potently to SMARCA2/4 and PB1(5) as determined by ITC and shows weak affinity for the highly conserved homologues PB1(2) and PB1(3).
SGC-SMARCA-BRDVIII (22) is cell active and it impairs the formation of adipocytes from 3T3-L1 fibroblasts by reducing the expression levels of adipocyte-related genes. The control compound SGC-BRDVIII-NC (35) is not active.
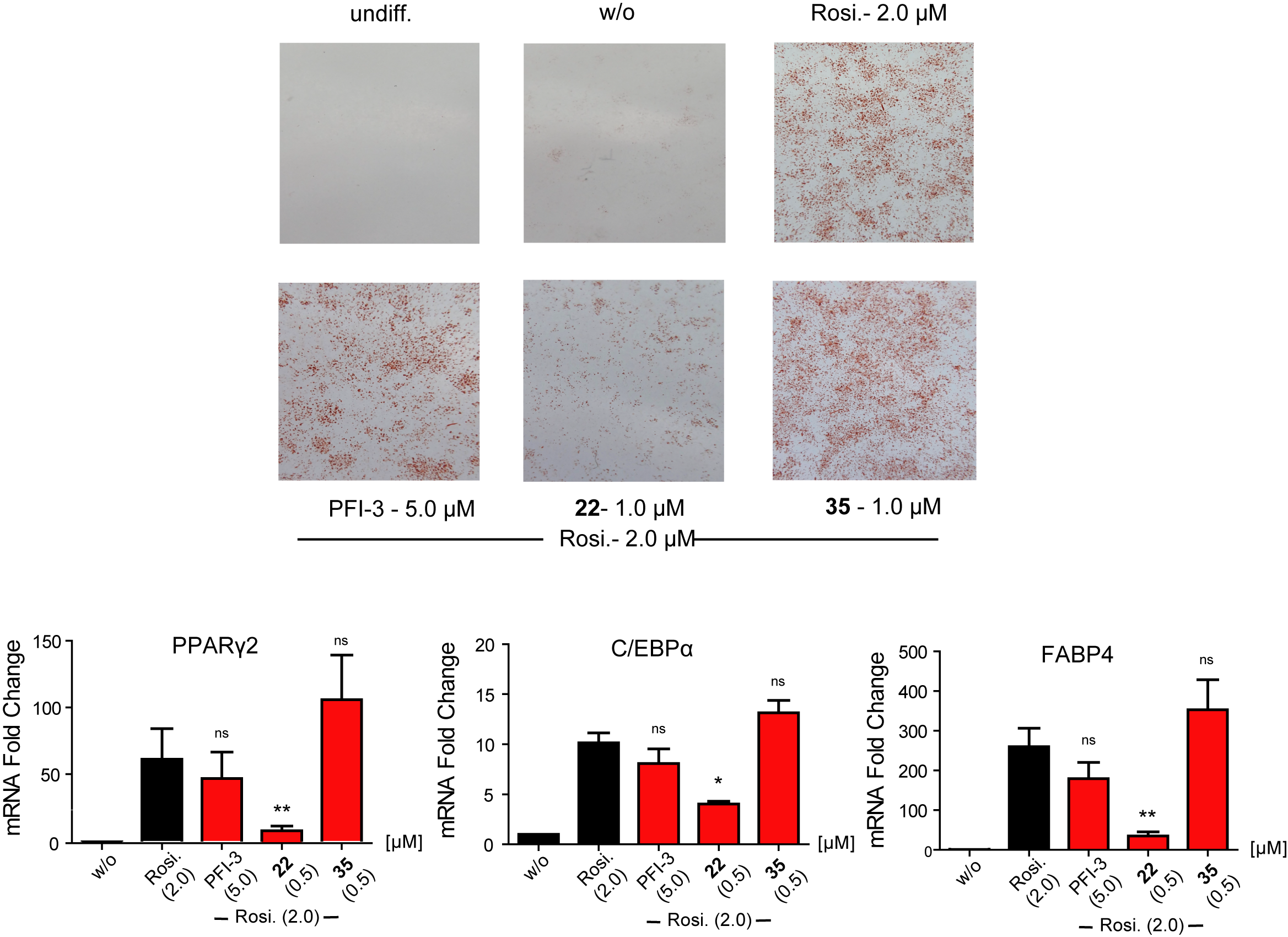
Figure 1: Cell based assay data for SGC-SMARCA-BRDVIII
SGC-SMARCA-BRDVIII is non-toxic and does not influence cell growth at a dose of 10 µM as indicated in the NCI-60 human tumor cell lines screen, and can therefore ideally used in cell differentiation assays.
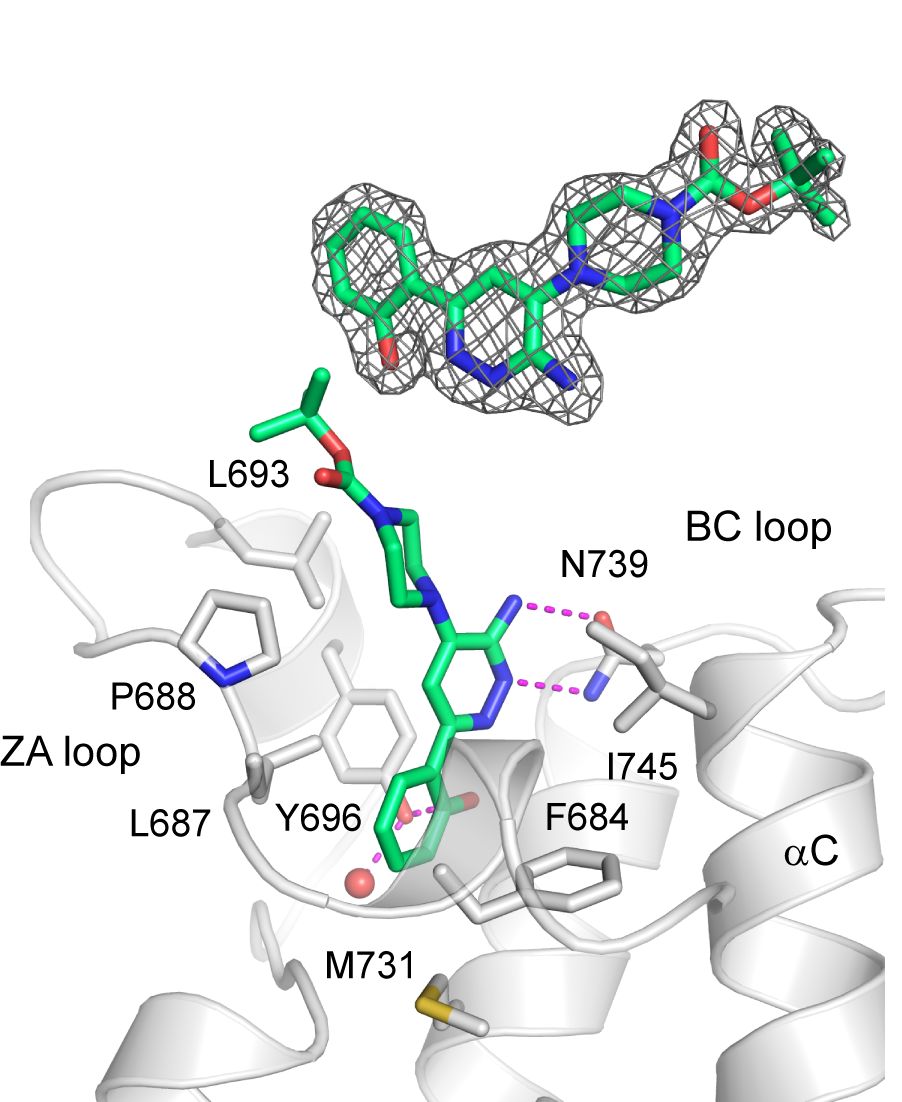
Binding mode of SGC-SMARCA-BRDVIII in complex with PB1(5)-BRD (PDB-ID 6ZS4). The inhibitor binds to the highly conserved Asn739 and Tyr696. The methylation of the hydroxy group in SGC-BRDVIII-NC blocks its binding to the bromodomain K(ac) binding pocket.