The probe is available from Tocris , Cayman Chemical and Sigma
| Probe |
 |
BAZ2-ICR |
Biology of the BAZ2 Bromodomains
BAZ2A/B belong to a family of ubiquitously expressed bromodomain containing proteins. Proteins of the BAZ (bromodomain adjacent zinc finger) family are characterized by a carboxy-terminal bromodomain adjacent to a PHD finger and a WACZ motif. In addition four other conserved motifs are typically found in the N-terminus of BAZ family members, namely the LH motif (a leucine-rich helical domain), the ZB2 motif (conserved between ZK783.4 and BAZ2 proteins) and the BAZ 1 and BAZ 2 motifs (conserved between all BAZ proteins, Acf, and ZK783.4).
Little is known about the biological function of BAZ2B but it has been suggested to regulate nucleosome mobilization by the ATP-dependent chromatin remodeling factor ISWI. Interaction of BAZ2B with ISWI is mediated by the BAZ1 motif [1]. The BAZ2B locus has also been identified as associated with sudden cardiac death [2]. The related BAZ2A (also called TIP5) protein plays a central role in the NoRC (nucleolar remodeling complex). BAZ2A is essential for heterochromatin formation leading to transcriptional silencing of certain rRNAs [3].
BAZ2-ICR
A collaboration between the SGC and Swen Hoelder’s group at the Institute of Cancer Research, London (ICR) has resulted in the discovery of BAZ2-ICR, a chemical probe for BAZ2A/B [4]. This is the second probe identified against this target, GSK2801, has previously been reported [5].
Co-crystal structure
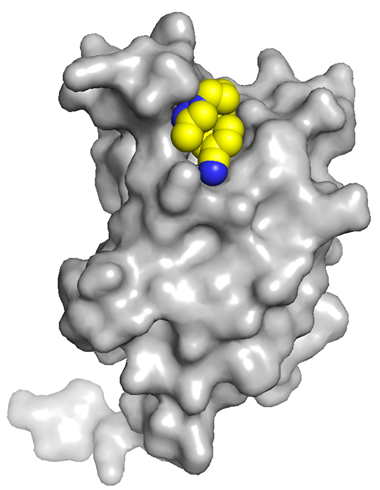
The co-crystal structure of BAZ2-ICR and BAZ2B has been solved (pdb id 4XUB)
Potency Against Target Family
Selectivity
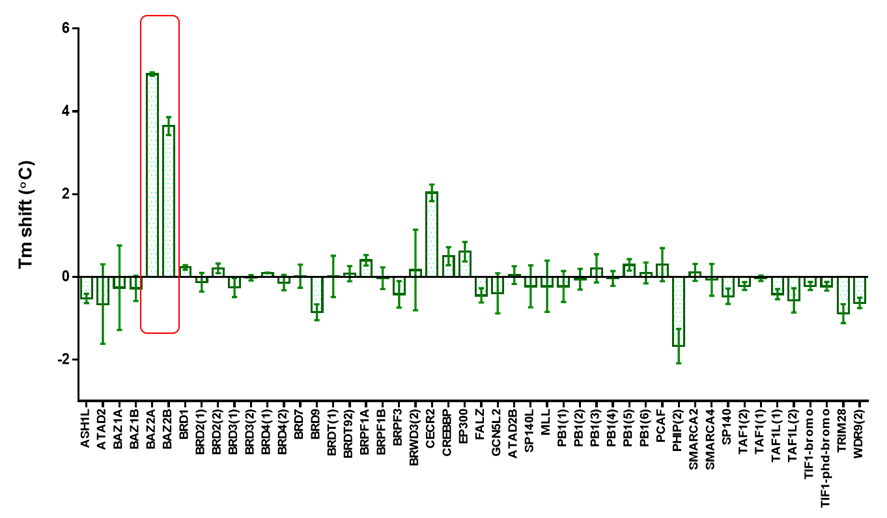
The inhibitor was screened at 10 μM concentration against 47 bromodomains using a temperature shift assay. The screened targets are labelled on the phylogenetic tree; targets that have not been screened are shown in grey. Temperature shifts are represented as spheres as indicated in the figure.
electivity Beyond Target Family
BAZ2-ICR (10 µm) displayed no activity against a panel of 55 receptors and ion channels
Cellular Activity
BAZ2-ICR shows accelerated FRAP recovery at 1 µM in the BAZ2A FRAP assay
| BAZ2-ICR |
 |
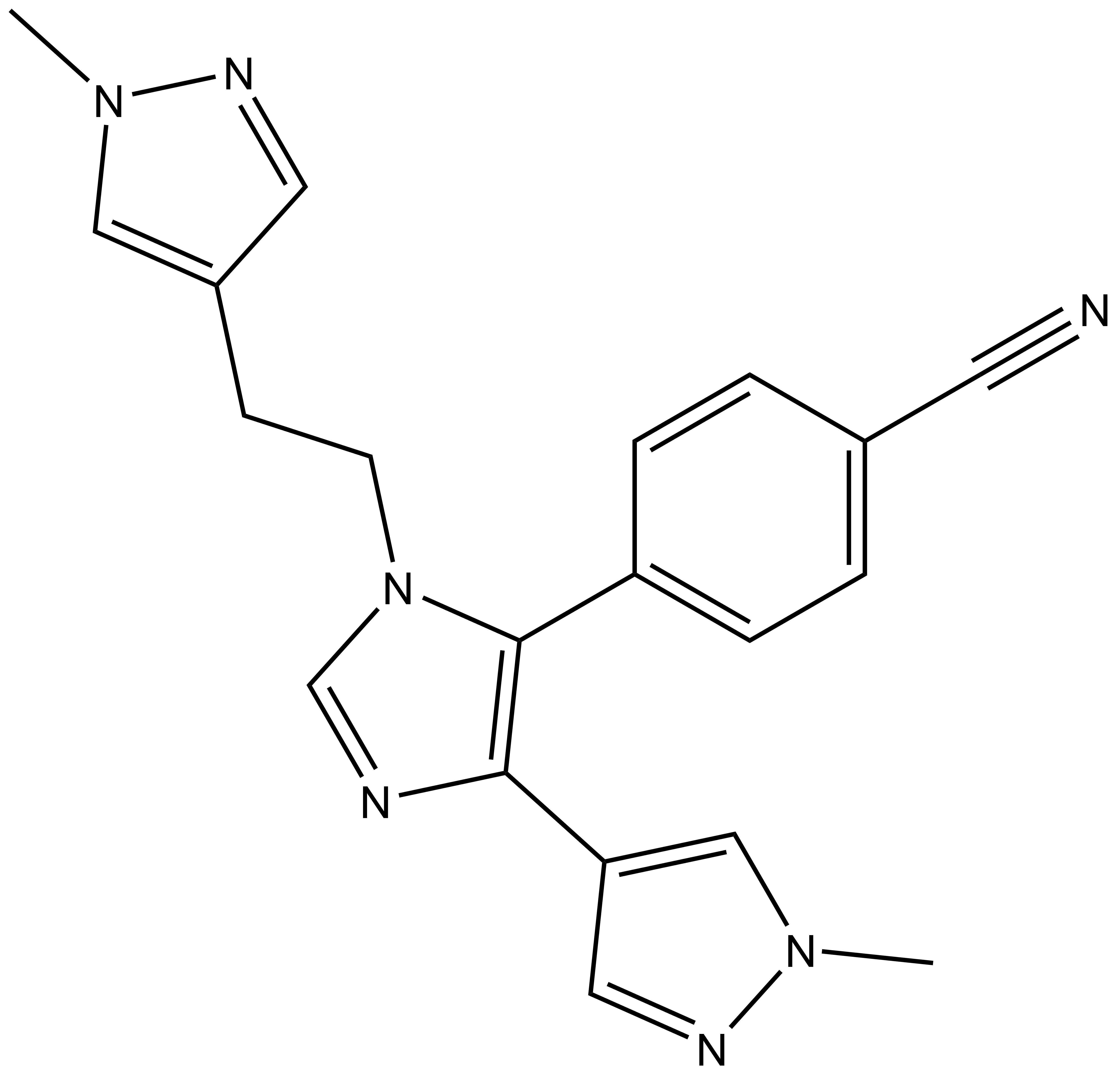
4-(4-(1-methyl-1H-pyrazol-3-yl)-1-(2-(1-methyl-1H-pyrazol-4-yl)ethyl)-1H-imidazol-5-yl)benzonitrile |
An inactive, negative control, compound for BAZ2-ICR has yet to be identified
| Physical and chemical properties | |
|---|---|
| Molecular weight | 357.42 |
| Molecular formula | C20H19N7 |
| IUPAC name | 4-(4-(1-methyl-1H-pyrazol-3-yl)-1-(2-(1-methyl-1H-pyrazol-4-yl)ethyl)-1H-imidazol-5-yl)benzonitrile |
| logP | 2.54 |
| PSA | 70.59 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 5 |
| No. of hydrogen bond acceptors | 7 |
| No. of hydrogen bond donors | 0 |
| Storage | Stable as solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | 25 mM in D2O |
Potency against Target
BAZ2-ICR has been shown by ITC to bind to BAZ2A with a KD of 109 nM (ITC) and to BAZ2B with a KD of 170 nM. The strong negative binding enthalpy possibly due to aromatic stacking interaction of pyrozole/phenyl
Selectivity Within Target family
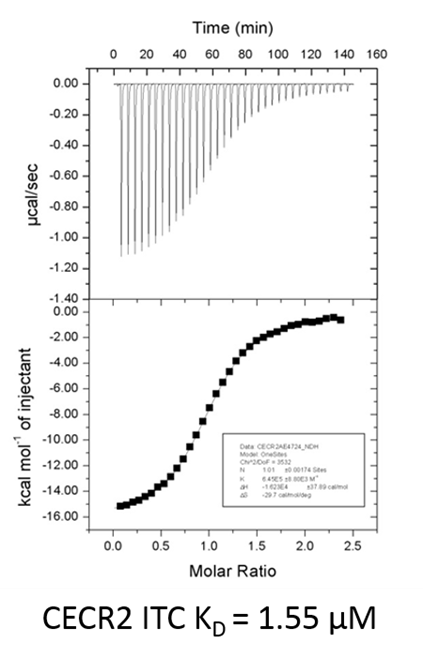
BAZ2-ICR showed significant thermal shifts of 5.2 and 3.8 °C for BAZ2A and BAZ2B respectively. BAZ2-ICR did not show significant thermal shifts against all other bromodomains, except for Cat Eye syndrome chromosome region, candidate 2 (CECR2), where a smaller but significant shift (ΔTm: 2.0 °C) was observed. The Kd of BAZ2-ICR binding to CERC2 was determined to be 1.55 μM by ITC, giving a 15-fold and 10-fold selectivity over BAZ2A and BAZ2B respectively.
Click the images to enlarge them
AlphaScreen Assay
To confirm selectivity over BRD4, BAZ2-ICR assay was studied in a BRD4 AlphaScreen assay, where it did not show significant inhibition up to 50 μM, translating in greater than 100-fold window.
Selectivity Beyond Target Family
BAZ2-ICR (10 µm) displayed no activity against a panel of 55 receptors and ion channels
FRAP
To investigate whether BAZ2-ICR can displace BAZ2 bromodomains from chromatin in living cells, we performed a fluorescence recovery after photobleaching (FRAP) assay utilizing GFP-tagged BAZ2A full length protein transfected into human osteosarcoma cells (U2OS). Mutant BAZ2A (N1873F) that does not bind acetylated lysine containing peptides and therefore mirrors the behaviour of inhibitor bound BAZ2A was used as a control. The histone deacetylase (HDAC) inhibitor suberoylanilide hydroxamic acid (SAHA) was used to increase overall levels of histone acetylation, resulting in a sufficient assay window to enable the measurement of differences in recovery time and demonstrating the acetylation dependence of the FRAP experiments. BAZ2-ICR at 1 μM reduced the recovery time of the wt construct to a level similar to the mutant, confirming that BAZ2-ICR inhibits BAZ2A in cells. Attempts to clone full length BAZ2B for this assay were unsuccessful.
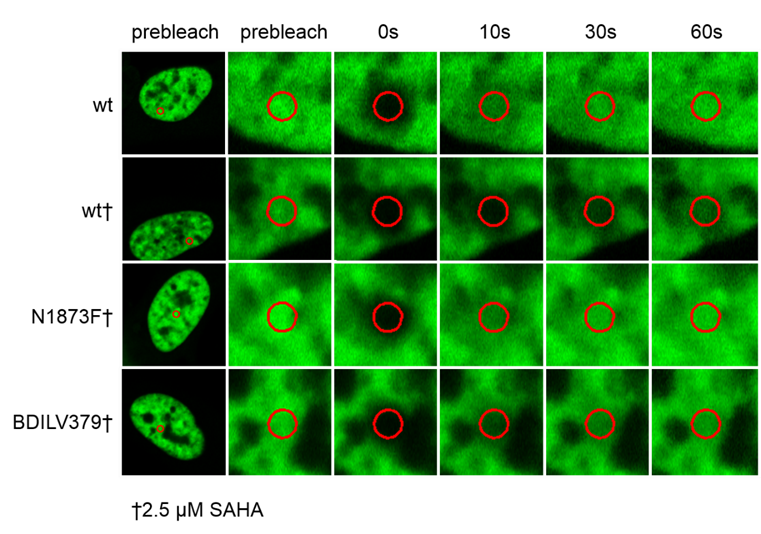
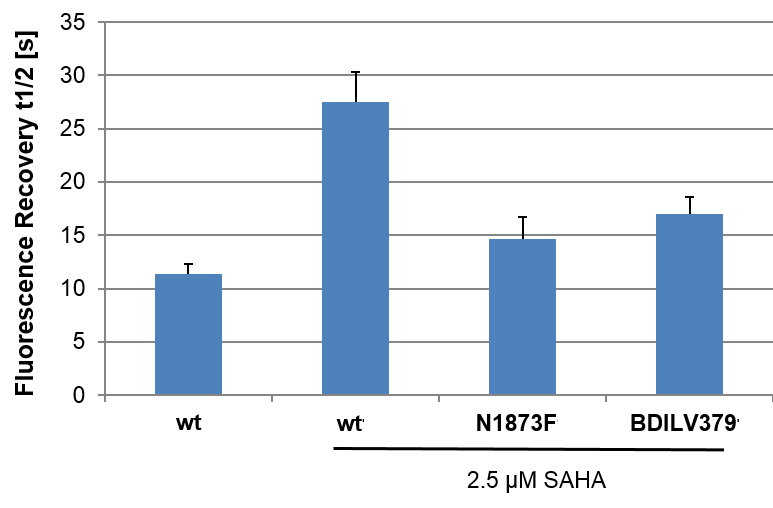
Click the images to enlarge them
To confirm selectivity in the cellular setting, a similar FRAP assay for BRD4 selectivity was performed. Cells were transfected with GFP-tagged BAZ2A or BRD4 and treated with 1 μM PFI-1 (a selective probe for BET Bromodomains) or BAZ2-ICR. Light bars depict assay with SAHA addition, and dark bars depict assays without SAHA addition (2.5 μM). Fluorescence recovery (t½) expressed as a percentage of the relevant wild-type control cells without inhibitor; the dotted line demarks the point equivalent to 100% of the relevant wild-type control cells without inhibitor. Error bars depict the standard error of the mean. Bars marked with an asterisk indicate a significant difference from controls (P < 0.05).
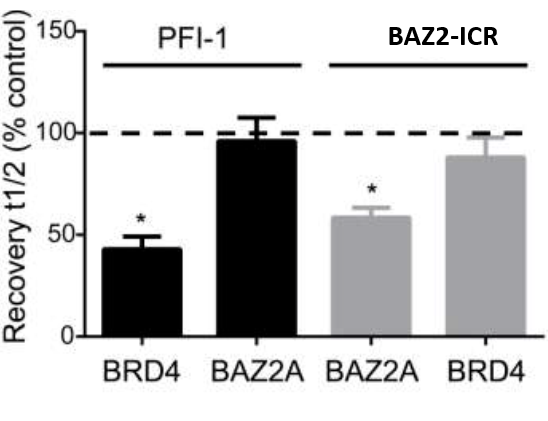
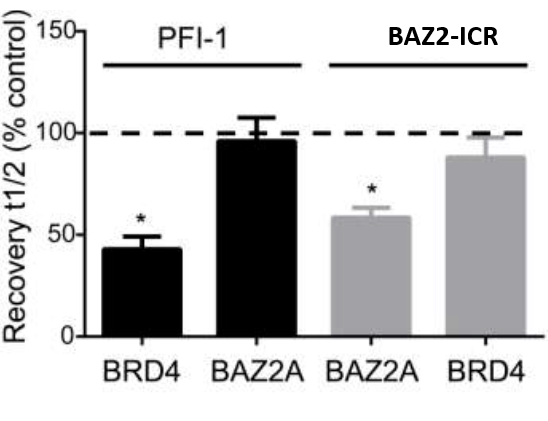
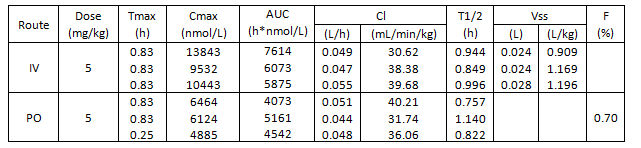
The physicochemical and mouse pharmacokinetic properties of BAZ2-ICR were assessed. BAZ2-ICR showed very high solubility (25 mM in D2O), a measured log D of 1.05, high stability in mouse microsomes, and permeation in the CaCo-2 model and thus a suitable profile for oral and intravenous gavage. We therefore performed a full mouse pharmacokinetic experiment. In agreement with the in vitro data, BAZ2-ICR showed 70% bioavailability and moderate clearance (∼50% of mouse liver blood flow) and volume of distribution, suggesting that BAZ2-ICR is suitable for modulating BAZ2A and BAZ2B in vivo.
3 mice were dosed IV and 3 mice PO. Key parameters and the curves are shown for each animal.
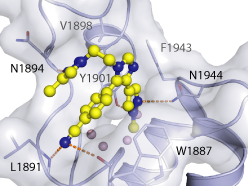
The co-crystal structure of BAZ2-ICR and BAZ2B has been solved (pdb id 4XUB)
The key interactions are illustrated below.
Isothermal Titration Calorimetry
Experiments were carried out on a VP-ITC titration microcalorimeter from MicroCalTM, LLC (Northampton, MA), at 15 °C while stirring at 295 rpm, in ITC buffer (50 mM HEPES, 150 mM NaCl). All titrations were conducted using an initial injection of 2 µl followed by 34 identical injections of 8 µl with a duration of 16 sec/injection and a spacing of 250 sec between injections. The heat of dilution was determined by independent titrations (protein into buffer) and was subtracted from the experimental data. MicroCalTM Origin software was used to calculate enthalpies of binding (ΔH) and binding constants (KB). Thermodynamic parameters were calculated (ΔG = ΔH - TΔS = -RTlnKB, where ΔG, ΔH and ΔS are the changes in free energy, enthalpy and entropy of binding respectively). In all cases a single binding site model was employed.
Differential Scanning Fluorimetry (DSF)
Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Stratagene). Proteins were buffered in 10 mM HEPES pH 7.5, 500 mM NaCl and assayed in a 96-well plate at a final concentration of 2 µM in 20 µl volume. Compounds were added at a final concentration of 10 µM. SYPRO Orange (Molecular Probes) was added as a fluorescence probe at a dilution of 1:1000. Excitation and emission filters for the SYPRO-Orange dye were set to 465 nm and 590 nm, respectively. The temperature was raised with a step of 3 °C per minute from 25 °C to 96 °C and fluorescence readings were taken at each interval. Data was analysed as previously reported [7]
CEREP assay
BAZ2-ICR (10 µm) was screened against a panel of 55 ligand receptors, ion channels and transports using an established and widely utilized commercial assay platform (ExpresSProfile; CEREP, Paris, FRANCE).
Fluorescence Recovery After Photobleaching (FRAP) Assay
FRAP studies were performed using U20S cells expressing full-length BAZ2A or BRD4 protein fused with an N-terminal eGFP as previously described [4]. Six hours after transfection 2.5uM SAHA (to increase global histone acetylation) was added and BAZ2-ICR was added 1 hour before imaging, which was carried out 24 hours after transfection. Percent inhibition was calculated between the DMSO treated (0%) and N1873F expressing mutant (100%)