This compound is available from Tocris, Sigma and Cayman Chemical
| Probe |
 |
Bromosporine (BSP) |
Bromodomains (BRDs) are protein interaction modules that read epigenetic marks recognizing ε-N-lysine acetylation motifs. The conserved BRD fold contains a deep, largely hydrophobic acetyllysine binding site, an attractive pocket for the development of small, pharmaceutically active molecules. BRDs have an important role in the targeting of chromatin-modifying enzymes to specific sites, including methyltransferases, HATs and transcription factors and regulate diverse biological processes from cell proliferation and differentiation to energy homeostasis and neurological processes.
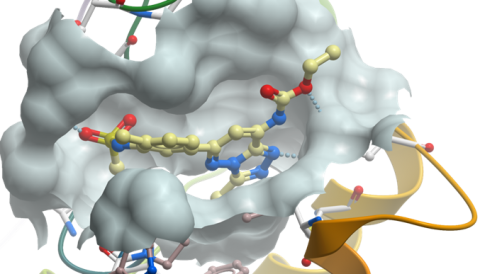
Co-crystal structure of Bromosporine with BRD4(1)
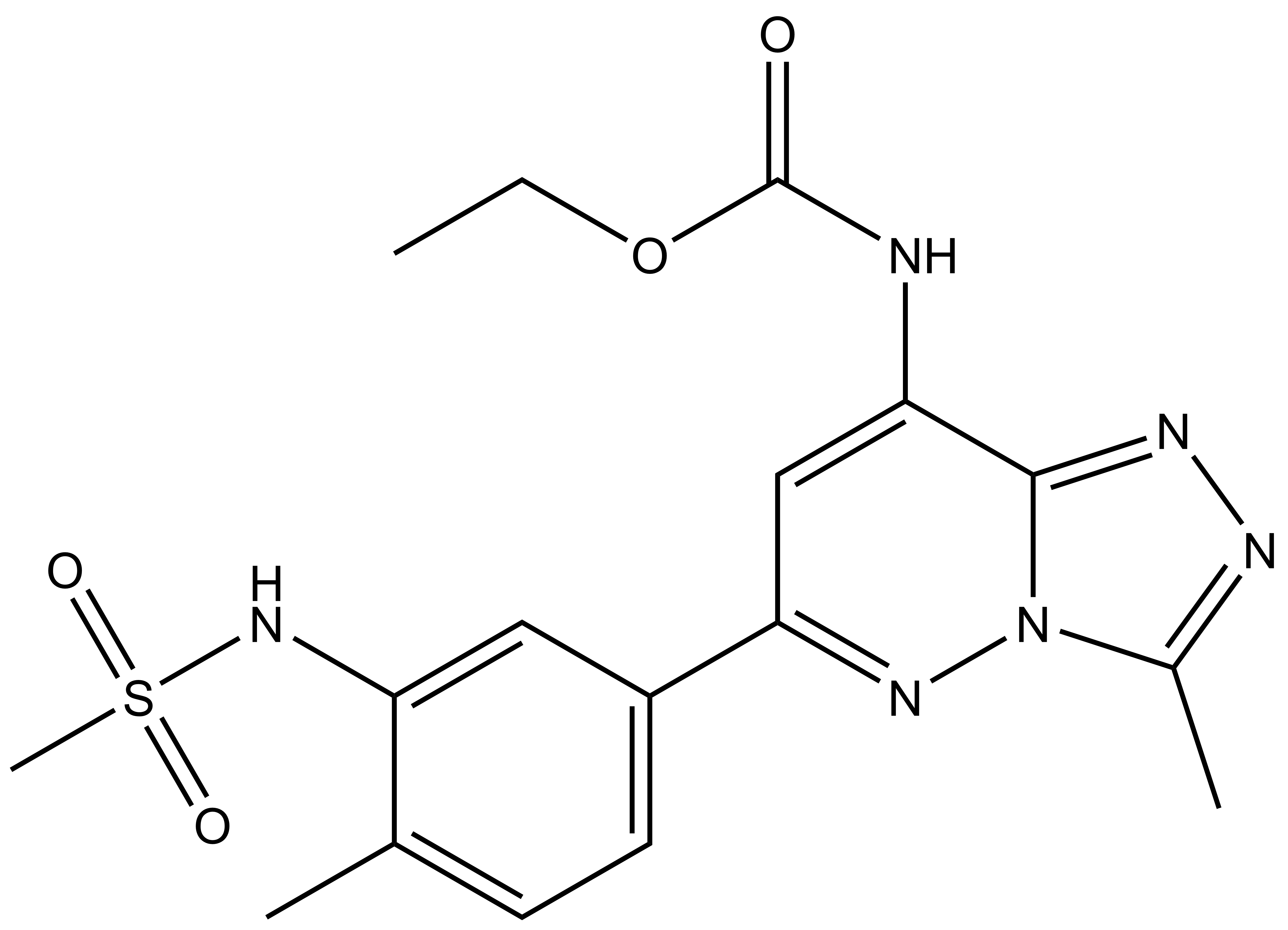
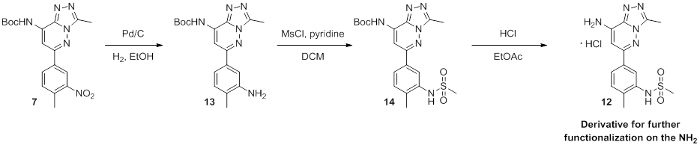
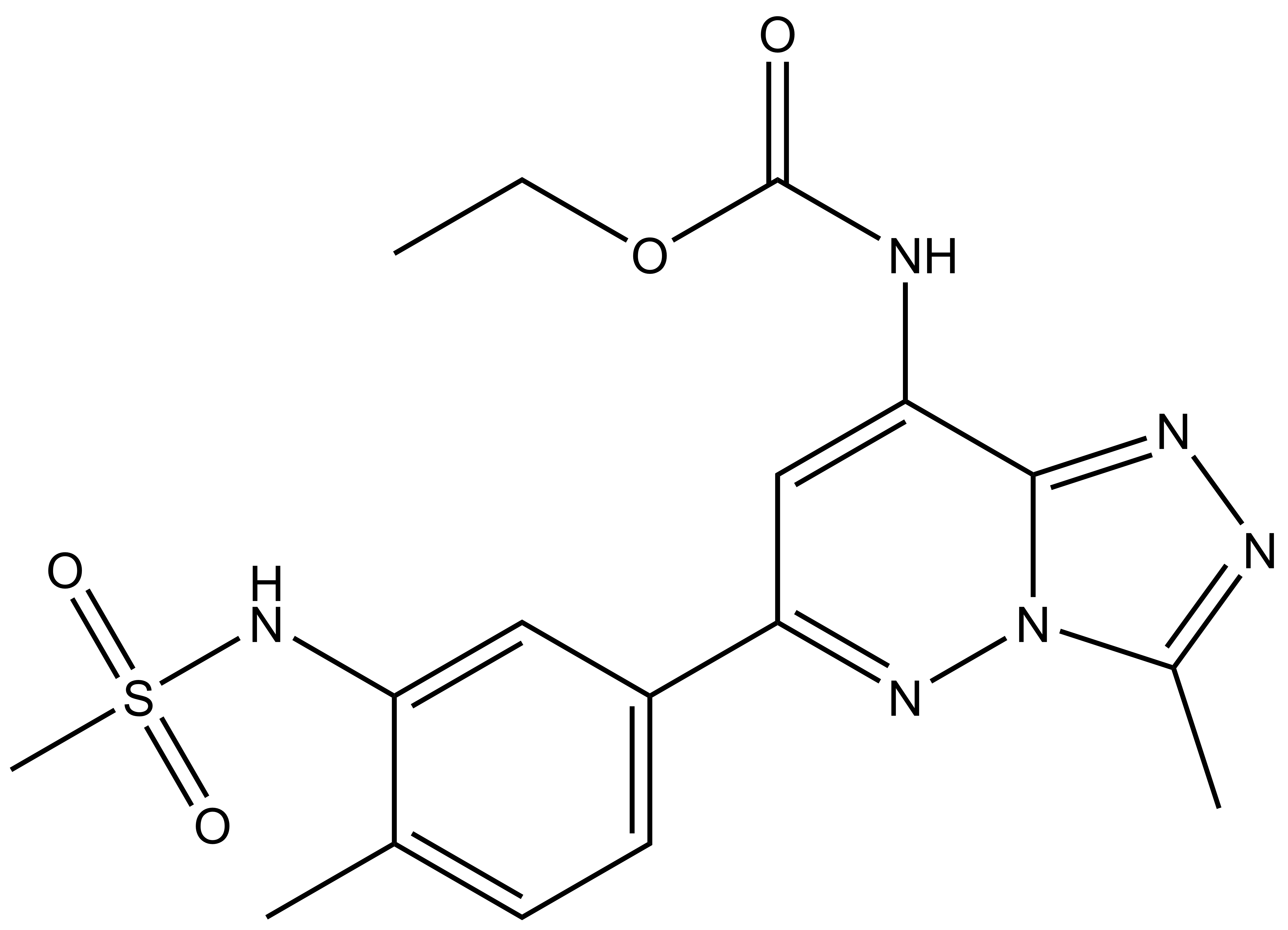
ethyl N-[6-(3-methanesulfonamido-4-methylphenyl)-3-methyl-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]carbamate
Click here to download SDF file
| Physical and chemical properties | |
|---|---|
| Molecular weight | 404.44 |
| Molecular formula | C17H20N6O4S |
| IUPAC Name | ethyl N-[6-(3-methanesulfonamido-4-methylphenyl)-3-methyl-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]carbamate |
| clogP | 0.78 |
| PSA | 105.64 |
| Storage | 2-8°C as powder. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | Soluble in DMSO at least up to 50mM |
CC1=NN=C2N1N=C(C3=CC=C(C)C(NS(=O)(C)=O)=C3)C=C2NC(OCC)=O
InChI=1S/C17H20N6O4S/c1-5-27-17(24)18-15-9-14(21-23-11(3)19-20-16(15)23)12-7-6-10(2)13(8-12)22-28(4,25)26/h6-9,22H,5H2,1-4H3,(H,18,24)
UYBRROMMFMPJAN-UHFFFAOYSA-N
Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Stratagene). Proteins were buffered in 10 mM HEPES pH 7.5, 500 mM NaCl and assayed in a 96-well plate at a final concentration of 2 µM in 20 µl volume. Bromosporine was added at a final concentration of 10 µM. SYPRO Orange (Molecular Probes) was added as a fluorescence probe at a dilution of 1:1000. Excitation and emission filters for the SYPRO-Orange dye were set to 465 nm and 590 nm, respectively. The temperature was raised with a step of 3 °C per minute from 25 °C to 96 °C and fluorescence readings were taken at each interval. The temperature dependence of the fluorescence during the protein denaturation process was approximated by the equation (displayed below) where ΔuG(T) is the difference in unfolding free energy between the folded and unfolded state, R is the gas constant and yF and yU are the fluorescence intensity of the probe in the presence of completely folded and unfolded protein respectively. The baselines of the denatured and native states were approximated by a linear fit. The observed temperature shifts, ΔTmobs, were recorded as the difference between the transition midpoints of sample and reference wells containing protein without ligand in the same plate and determined by non-linear least squares fit.
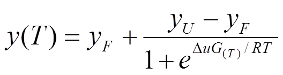
| Selectivity | |
|---|---|
| Bromodomain | Thermal melt |
| 10 µM ΔTm | |
| BRD2(1) | 4.4 |
| BRD2(2) | 5.9 |
| BRD3(1) | 5.3 |
| BRD3(2) | 5.9 |
| BRD4(1) | 6.9 |
| BRD4(2) | 6.2 |
| BRDT(1) | 7.3 |
| BRDT(2) | 5.2 |
| CECR2 | 8.3 |
| PB1(5) | 0.4 |
| TAF1(1) | 1.3 |
| TAF1(2) | 5.2 |
| TAF1L(1) | 0.9 |
| TAF1L(2) | 0.9 |
| BAZ2A | 1.2 |
| TIF1α | 0.4 |
| ATAD2 | -0.2 |
| BRD9 | 3.9 |
| CREBBP | 3.4 |
| Potency in Cells | |
|---|---|
| BRD4 | Accelerated FRAP recovery at 1 µM |
| CREBBP | Accelerated FRAP recovery at 1 µM |
| TIF1α | Inactive at 10 µM |
| BAZ2A | Inactive at 10 µM |
| SMARCA2 | Inactive at 10 µM |
Bromosporine shows moderate cytotoxicity in HeLa cells at 18 µM
Sarah Picaud, Katharina Leonards, Jean-Philippe Lambert, Oliver Dovey, Christopher Wells, Oleg Fedorov, Octovia Monteiro, Takao Fujisawa, Chen-Yi Wang, Hannah Lingard, Cynthia Tallant, Nikzad Nikbin, Lucie Guetzoyan, Richard Ingham, Steven V. Ley, Paul Brennan, Susanne Muller, Anastasia Samsonova, Anne-Claude Gingras, Juerg Schwaller, George Vassiliou, Stefan Knapp and Panagis Filippakopoulos. Science Advances 12 Oct 2016: Vol. 2, no. 10, e1600760. DOI: 10.1126/sciadv.1600760
Scheme 1
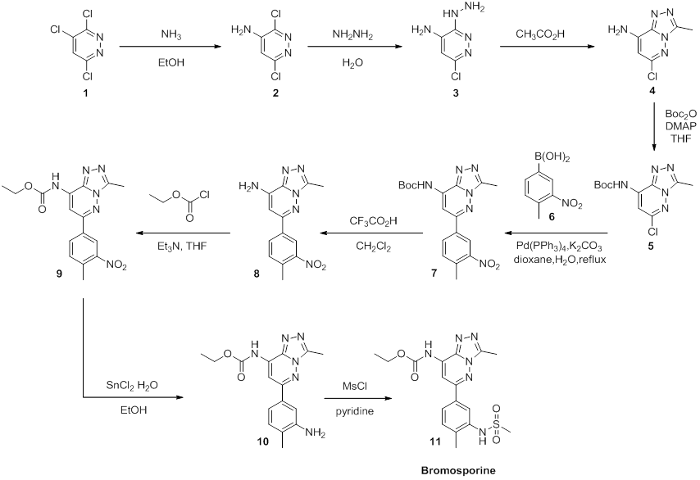
Scheme 2