| Probe | Negative control | |
 |
|  |
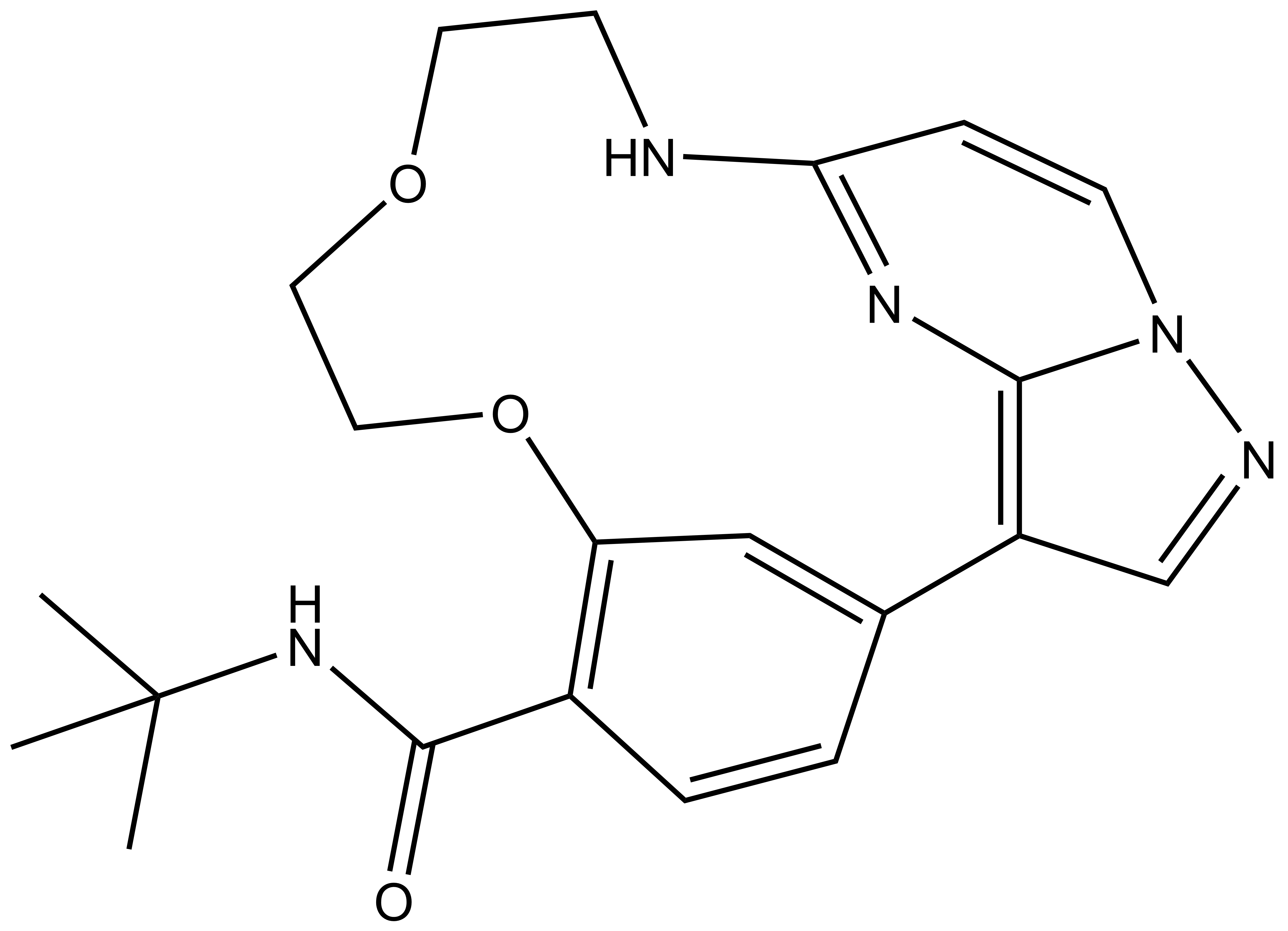
CK156 |
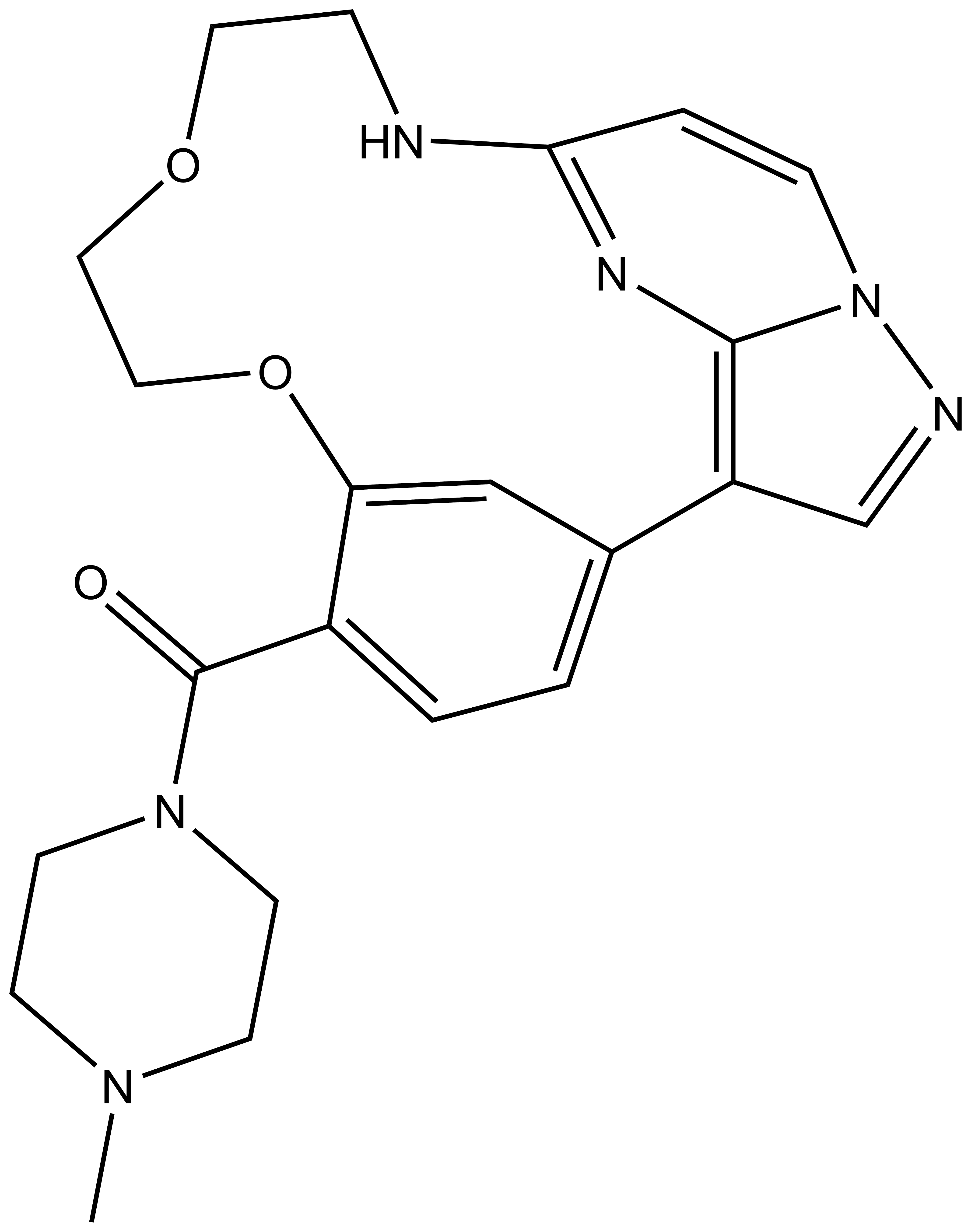
| CKJB71 |
DAP Kinase-Related Apoptosis-Inducing Protein Kinase 1 (DRAK1) is part of the DAPK (death-associated protein kinases) family, which comprises of five members (DAPK1–3 and DRAK2). Both, DRAK´s (DRAK1 & 2) belong to the so-called dark kinome and their cellular functions are largely unknown [1–3]. However, recent findings indicate that DRAK1 might play a role in different cancers such as glioblastoma multiforme (GBM) [4], head and neck squamous cell carcinoma (HNSCC) [5] or in testicular cancer [6]. In addition, DRAK1 has been identified to be a direct target gene of p53, and vice versa DRAK1 might regulate the transcriptional activity of p53 [6, 7]. More recently, Park et al demonstrated that DRAK1 might also act as a negative regulator of TRAF6, which is a central player in inflammatory signalling pathway in cervical cancer [8]. However, the precise role of DRAK1 in different cancers remains elusive and chemical tools are urgently needed to determine the role of DRAK1 in these diseases.
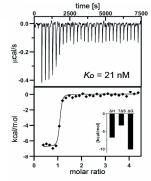
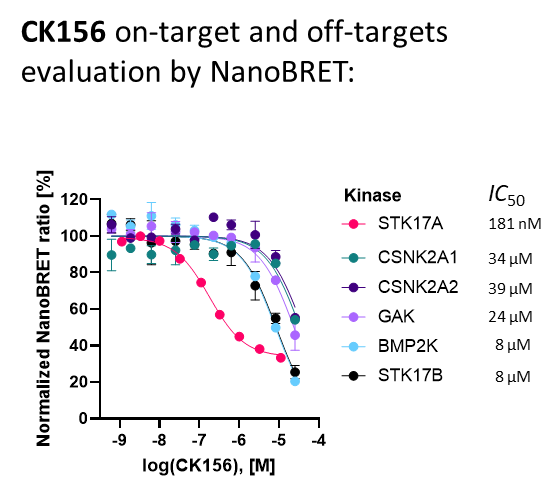
SGC has developed CK156, a potent and selective DRAK1 inhibitor with an IC50 of 49 nM for DRAK1 determined by a radiometric assay and a KD of 21 nM determined by ITC. Cellular activity was examined by NanoBRET and CK156 revealed an IC50 of 181 nM on DRAK1. The chemical probe (CK156) is accompanied by a negative control (CKJB71), which is structurally closely related to the probe molecule.
Potency Against Target Family
| Kinase | 33PanQinase IC50 (nM) |
| DRAK1 | 49 |
ITC: KD = 21 nM
Selectivity
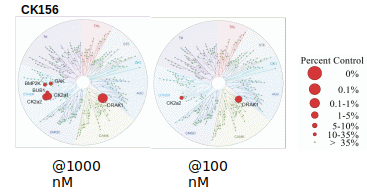
CK156 has been shown to be selective in an in vitro kinase panel from DiscoverX (scanMAX®) against 468 Kinases followed by cellular NanoBRET assays.
Dosage
To minimize the chance of any unspecific cytotoxicity, we recommend a concentration of no higher than 5 µM for cell-based assays.
Cellular Activity
CK156 displayed an IC50 of 181 nM in NanoBRETTM assay.
| Probe | Negative control | |
 |
|  |
CK156 |
| CKJB71 |
Physical and chemical properties CK156 | |
| Molecular weight | 395.46
|
| Molecular formula | C21H25N5O3 |
| IUPAC name | N-tert-butyl-7,10-dioxa-13,17,18,21-tetraazatetracyclo[12.5.2.1²,⁶.0¹⁷,²⁰]docosa-1(20),2,4,6(22),14(21),15,18-heptaene-5-carboxamide |
| logP | 1.89 |
| PSA | 89.78 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 2 |
| No. of hydrogen bond acceptors | 8 |
| No. of hydrogen bond donors | 2 |
| Storage | stable as solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | soluble in DMSO in a concentration of 10 mM |
SMILES: O=C(C(C=C1)=C(OCCOCCNC2=N3)C=C1C4=C3N(C=C2)N=C4)NC(C)(C)C
InChI: InChI=1S/C21H25N5O3/c1-21(2,3)25-20(27)15-5-4-14-12-17(15)29-11-10-28-9-7-22-18-6-8-26-19(24-18)16(14)13-23-26/h4-6,8,12-13H,7,9-11H2,1-3H3,(H,22,24)(H,25,27)
InChIKey: YCFFONJEHNSECY-UHFFFAOYSA-N
Physical and chemical properties CKJB71 | |
| Molecular weight | 422.49 |
| Molecular formula | C22H26N6O3 |
| IUPAC name | 5-(4-methylpiperazine-1-carbonyl)-7,10-dioxa-13,17,18,21-tetraazatetracyclo[12.5.2.1²,⁶.0¹⁷,²⁰]docosa-1(20),2,4,6(22),14(21),15,18-heptaene |
| logP | 1.00 |
| PSA | 84.23 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 1 |
| No. of hydrogen bond acceptors | 9 |
| No. of hydrogen bond donors | 1 |
| Storage | stable as solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | soluble in DMSO in a concentration of 10 mM |
SMILES: O=C(C1=CC=C2C=C1OCCOCCNC3=NC4=C2C=NN4C=C3)N5CCN(CC5)C
InChI: InChI=1S/C22H26N6O3/c1-26-7-9-27(10-8-26)22(29)17-3-2-16-14-19(17)31-13-12-30-11-5-23-20-4-6-28-21(25-20)18(16)15-24-28/h2-4,6,14-15H,5,7-13H2,1H3,(H,23,25)
InChIKey:
DDXOMWWURGFLKK-UHFFFAOYSA-N
Selectivity profile of CK156 was determined with the scanMAX® assay from DiscoverX at 1000 and 100 nM and on- and off targets were evaluated with in vitro IC50 with the 33PanQinase activity assay from ProQinase and in cellulo IC50 with NanoBRET assay.
Kinase | Percent of control(%) @ 1000 nM | 33PanQinase IC50 (nM) | NanoBRET IC50 (nM) |
| DRAK1 | 0.3 | 49 | 181 |
| CSNK2A2 | 1.5 | 380 | 39000 |
| CSNK2A1 | 3.9 | 950 | 34000 |
| GAK | 23 | n.d. | 24000 |
| BIKE | 25 | n.d. | 8000 |
| JAK3(JH1domain-catalytic) | 28 | n.d. | n.d |
| RIOK3 | 28 | n.d. | n.d. |
| BUB1 | 30 | n.d. | n.d. |
| ERK8 | 33 | n.d. | n.d |
| DRAK2 | 39 | n.d. | 8000 |
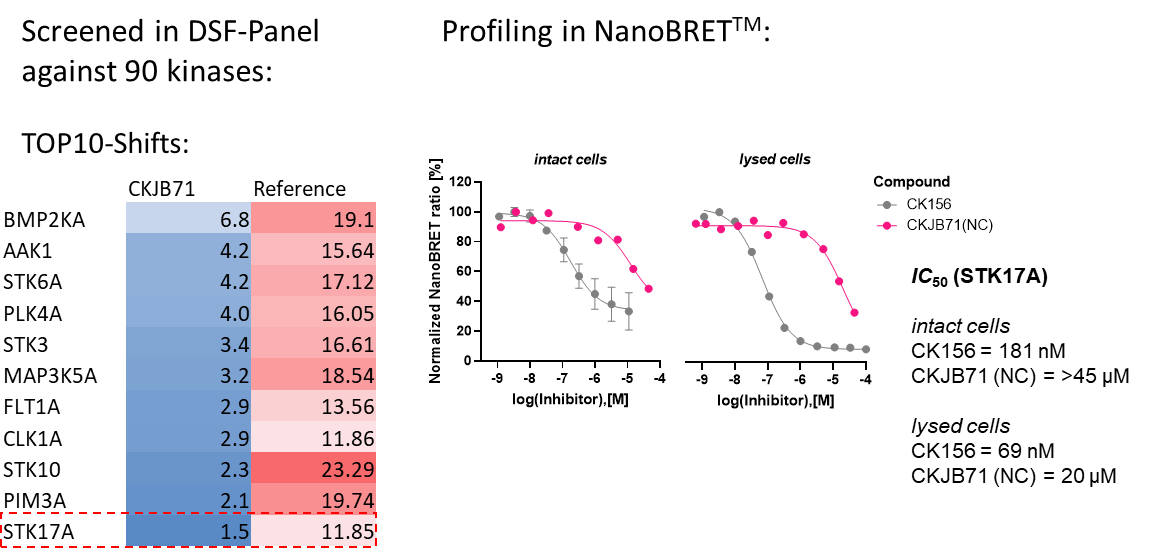
The negative control CKJB71 showed no activity in a DSF assay for 90 kinases and no on-target activity determined by NanoBRET in intact cells and in lysed cells.
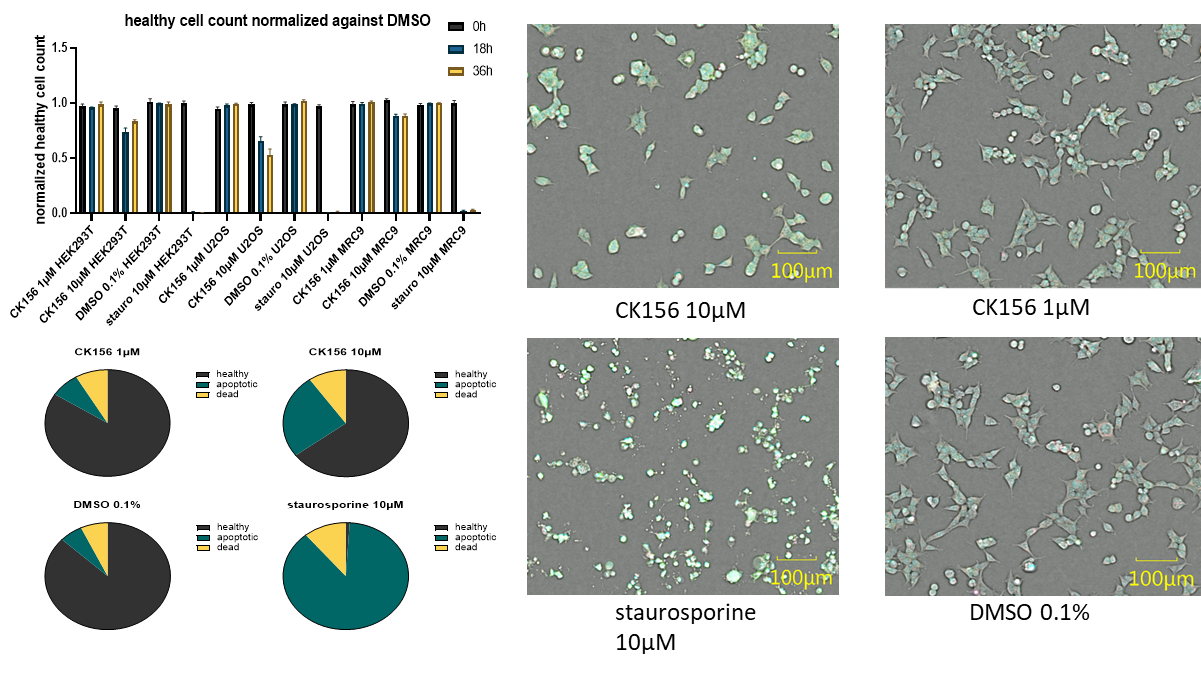
CK156 displayed no general cytotoxicity in three different cell lines (HEK293T, U2OS and MRC9-Fibroblasts) at 1µM and only slight toxicity at 10 µM. The cells were analysed after 18 and 36h.
[1] Berginski ME, Moret N, Liu C, Goldfarb D, Sorger PK, G. S. The Dark Kinase Knowledgebase: An Online Compendium of Knowledge and Experimental Results of Understudied Kinases. Nucleic Acids Res. 2020. https://doi.org/10.1093/nar/gkaa853.
[2] Farag, A. K.; Roh, E. J. Death-Associated Protein Kinase (DAPK) Family Modulators: Current and Future Therapeutic Outcomes. Medicinal Research Reviews. John Wiley and Sons Inc. January 1, 2019, pp 349–385. https://doi.org/10.1002/med.21518.
[3] Bialik, S.; Kimchi, A. The Death-Associated Protein Kinases: Structure, Function, and Beyond. Annual Review of Biochemistry. Annu Rev Biochem 2006, pp 189–210. https://doi.org/10.1146/annurev.biochem.75.103004.142615.
[4] Mao, P.; Hever-Jardine, M. P.; Rahme, G. J.; Yang, E.; Tam, J.; Kodali, A.; Biswal, B.; Fadul, C. E.; Gaur, A.; Israel, M. A.; Spinella, M. J. Serine/Threonine Kinase 17A Is a Novel Candidate for Therapeutic Targeting in Glioblastoma. PLoS One 2013, 8 (11). https://doi.org/10.1371/journal.pone.0081803.
[5] Park, Y.; Kim, W.; Lee, J. M.; Park, J.; Cho, J. K.; Pang, K.; Lee, J.; Kim, D.; Park, S. W.; Yang, K. M.; Kim, S. J. Cytoplasmic DRAK1 Overexpressed in Head and Neck Cancers Inhibits TGF-Β1 Tumor Suppressor Activity by Binding to Smad3 to Interrupt Its Complex Formation with Smad4. Oncogene 2015, 34 (39), 5037–5045. https://doi.org/10.1038/onc.2014.423.
[6] Mao, P.; Hever, M. P.; Niemaszyk, L. M.; Haghkerdar, J. M.; Yanco, E. G.; Desai, D.; Beyrouthy, M. J.; Kerley-Hamilton, J. S.; Freemantle, S. J.; Spinella, M. J. Serine/Threonine Kinase 17A Is a Novel P53 Target Gene and Modulator of Cisplatin Toxicity and Reactive Oxygen Species in Testicular Cancer Cells. J. Biol. Chem. 2011, 286 (22), 19381–19391. https://doi.org/10.1074/jbc.M111.218040.
[7] Oue, Y.; Murakami, S.; Isshiki, K.; Tsuji, A.; Yuasa, K. Intracellular Localization and Binding Partners of Death Associated Protein Kinase-Related Apoptosis-Inducing Protein Kinase 1. Biochem. Biophys. Res. Commun. 2018, 496 (4), 1222–1228. https://doi.org/10.1016/j.bbrc.2018.01.175.
[8] Park Y, Pang K, Park J, Hong E, Lee J, Ooshima A, Kim HS, Cho JH, Han Y, Lee C, Song YS, Park KS, Yang KM, Kim SJ. Destablilization of TRAF6 by DRAK1 Suppresses Tumor Growth and Metastasis in Cervical Cancer Cells. Cancer Res. 2020 Jun 15;80(12):2537-2549. doi: 10.1158/0008-5472.CAN-19-3428.