The probe GSK-J1 is available from Tocris, Cayman Chemical (sodiumsalt) and Sigma .
The less-active control GSK-J2 is available from Tocris, Cayman Chemical (sodiumsalt) and Sigma (sodiumsalt).
The pro-drug GSK-J4 is available from Tocris, Cayman Chemical (hydrochloride) and Sigma.
The pro-drug GSK-J5 is available from Tocris (hydrochloride), Cayman Chemical (hydrochloride) and Sigma.
| Probe | Prodrug of GSK-J1 for cellular use | Less-active control | Prodrug of GSK-J2 for cellular use | |||
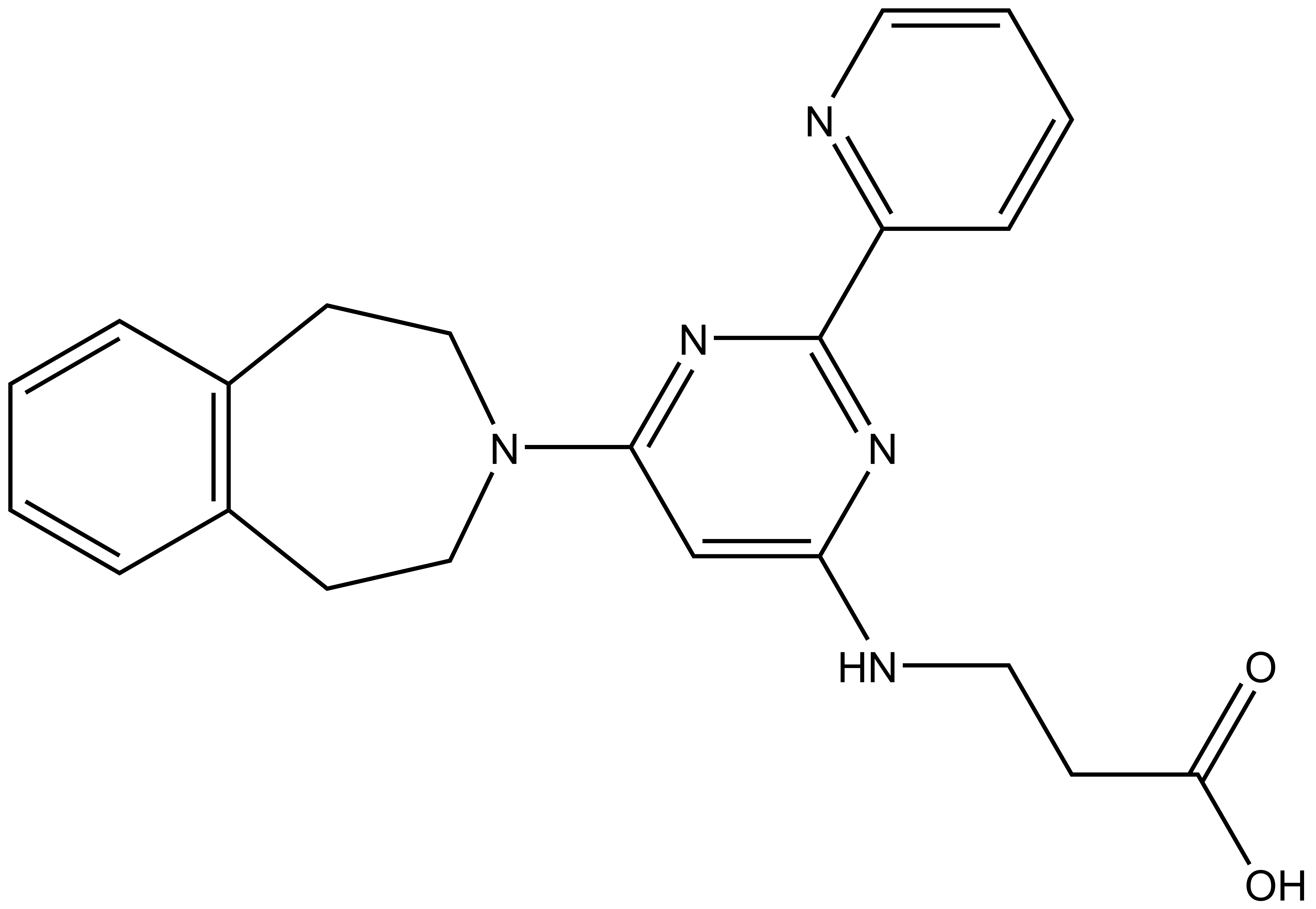 | 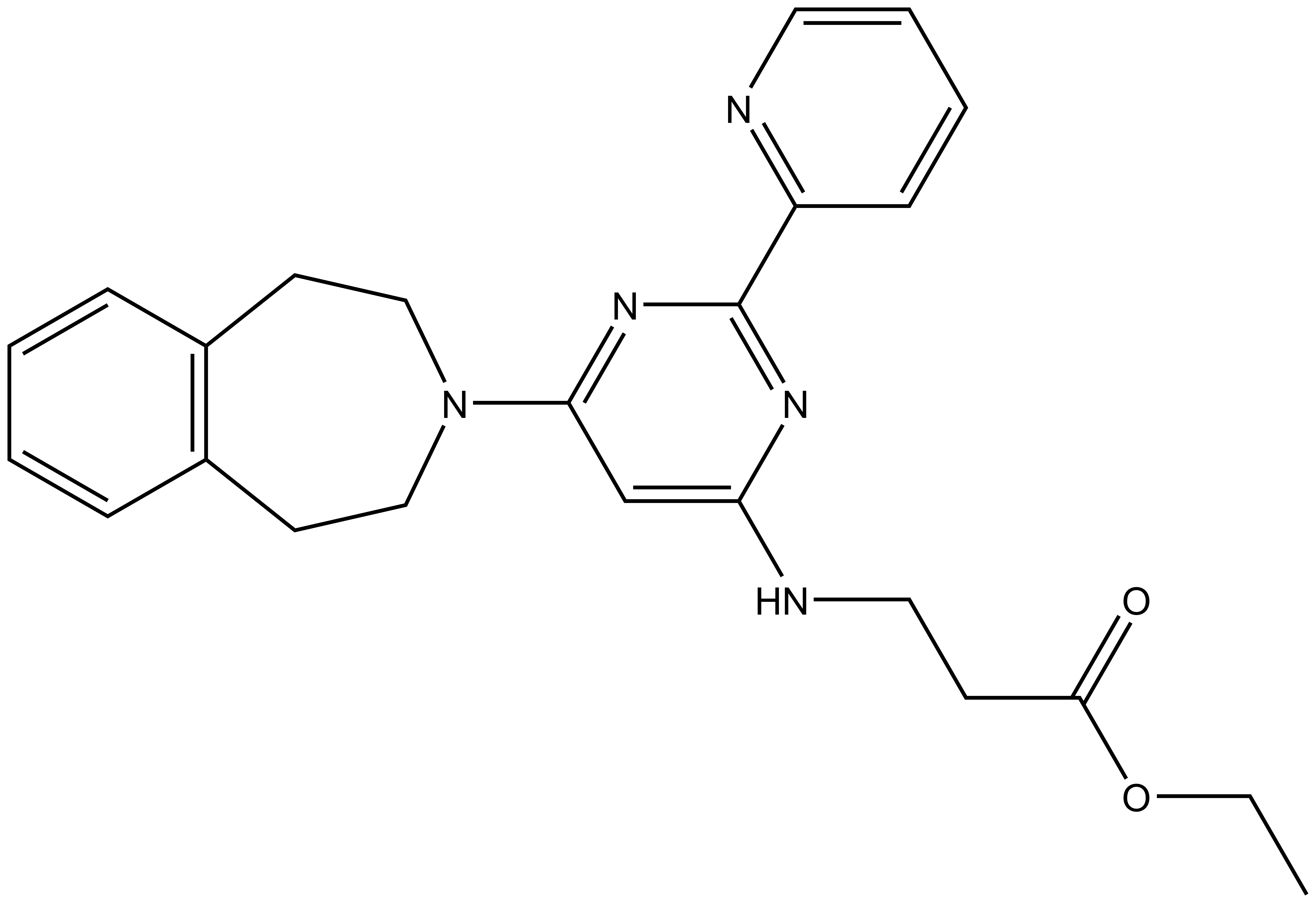 | 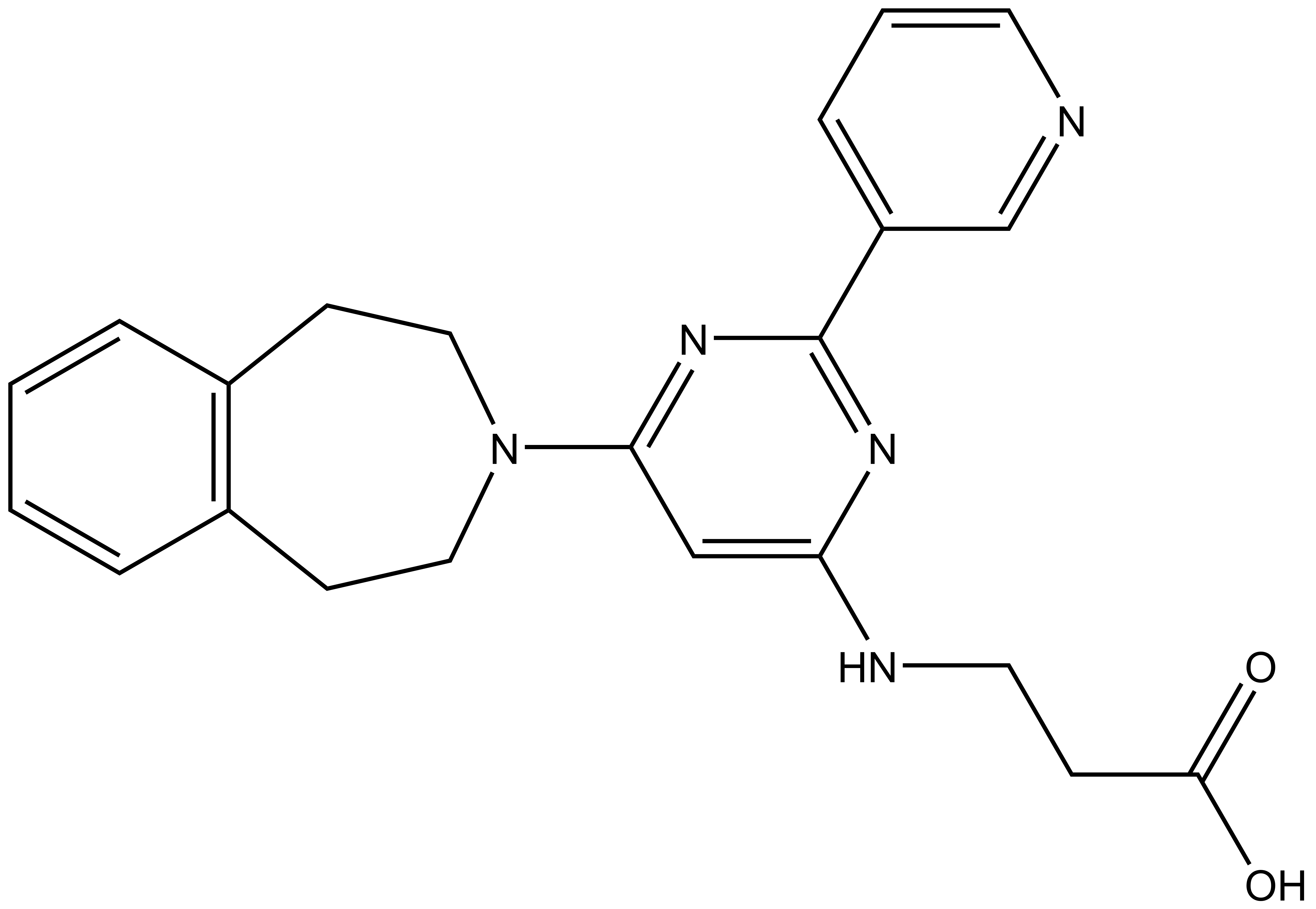 | 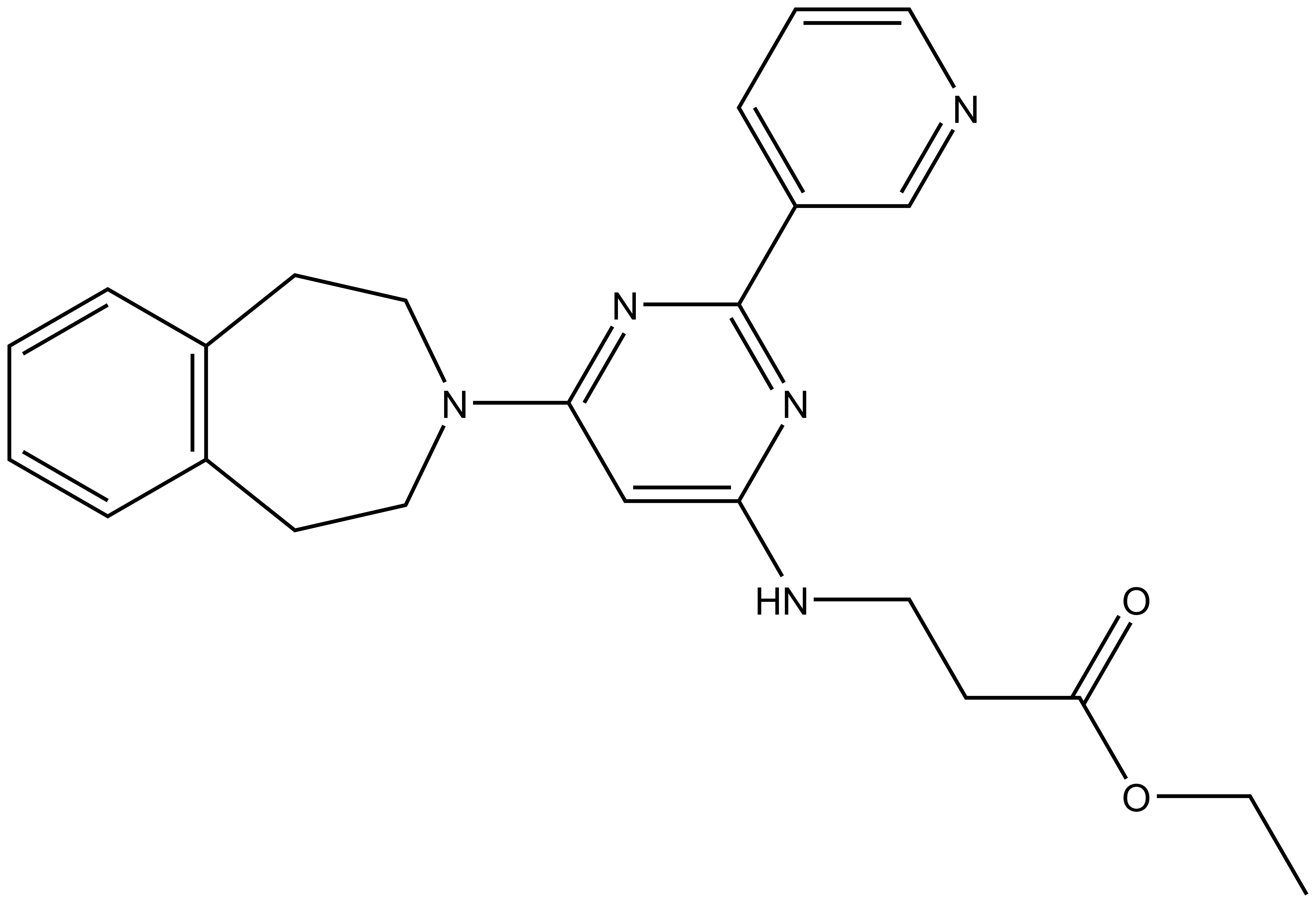 | |||
GSK-J1 | GSK-J4 | GSK-J2 | GSK-J5 |
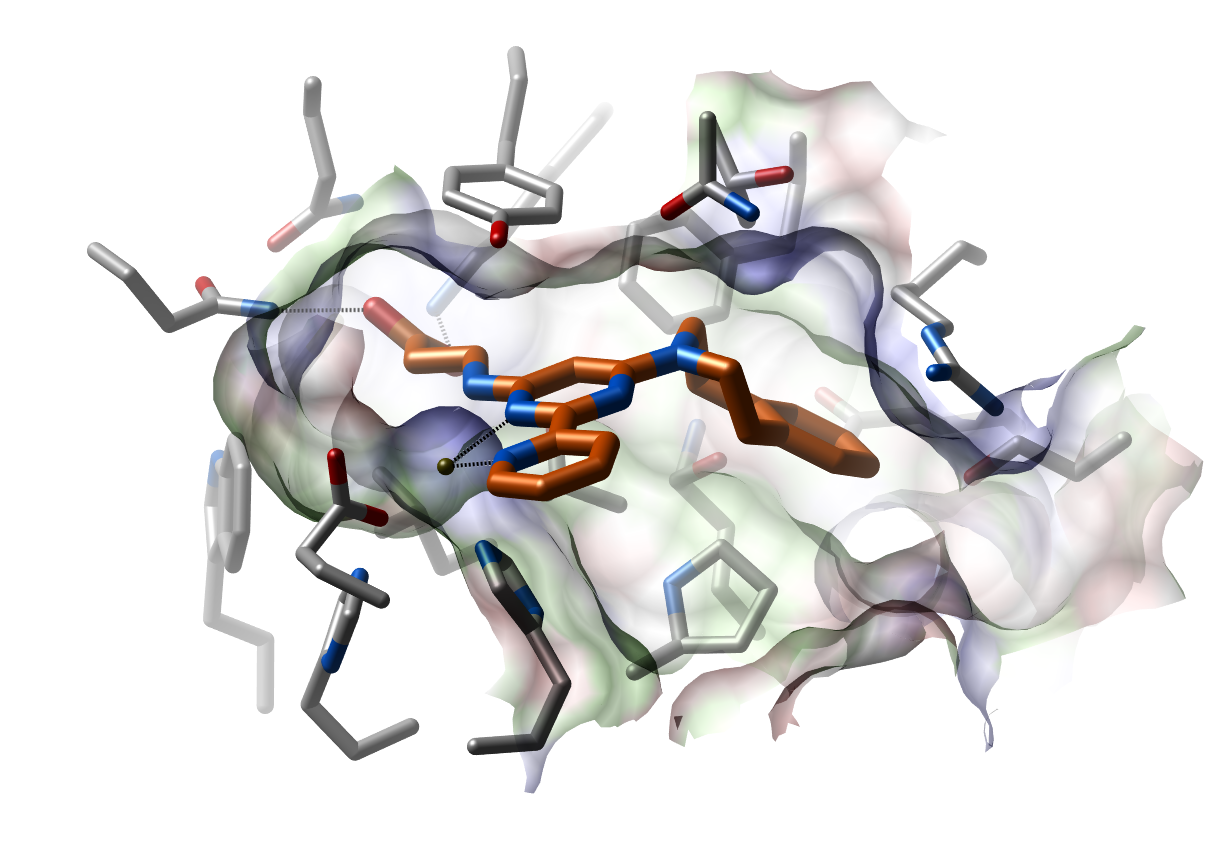
Co-crystal structure of JMJD3 in complex with GSKJ1 (PDB 4ASK). Residues within in the catalytic pocket of JMJD3 are shown as silver sticks, GSK-J1 is shown in orange sticks and Co2+ , that replaces the endogenous Fe2+ , is shown as a yellow sphere. (Click to enlarge)
Details of this study have been published in Nature - A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response.
Methylated lysine residues of histone 3 lysine 27 (H3K27me) play important roles in regulating gene activity by silencing through the polycomb-repressive complex (PRC1 or PRC2). This epigenetic mark can be demethylated by lysine demethylase (KDM) enzymes UTX and JmjD3 in humans and play important roles in cellular differentiation, development and cancer.
Here we present in collaboration with GlaxoSmithKline the first selective and potent histone demethylase inhibitor (GSK-J1) that has significant activity (IC50 60 nM for human JmjD3) in vitro and in cells using an ester derivative (GSK-J4: 1 µM < IC50 < 10 µM; e.g. 9 µM in primary human macrophages). The pyridine regio-isomer GSK-J2 displays significantly less on-target activity (IC50 > 100 µM for human JmjD3) and thus can be used as control for target effects in vitro, and as ester derivative (GSK-J5) in cells. Recent data has shown that GSK-J1 also shows some activity (IC50 950 nM for Jarid1b, IC50 1.76 uM for Jarid1c) against H3K4me3/2/1 demethylases.
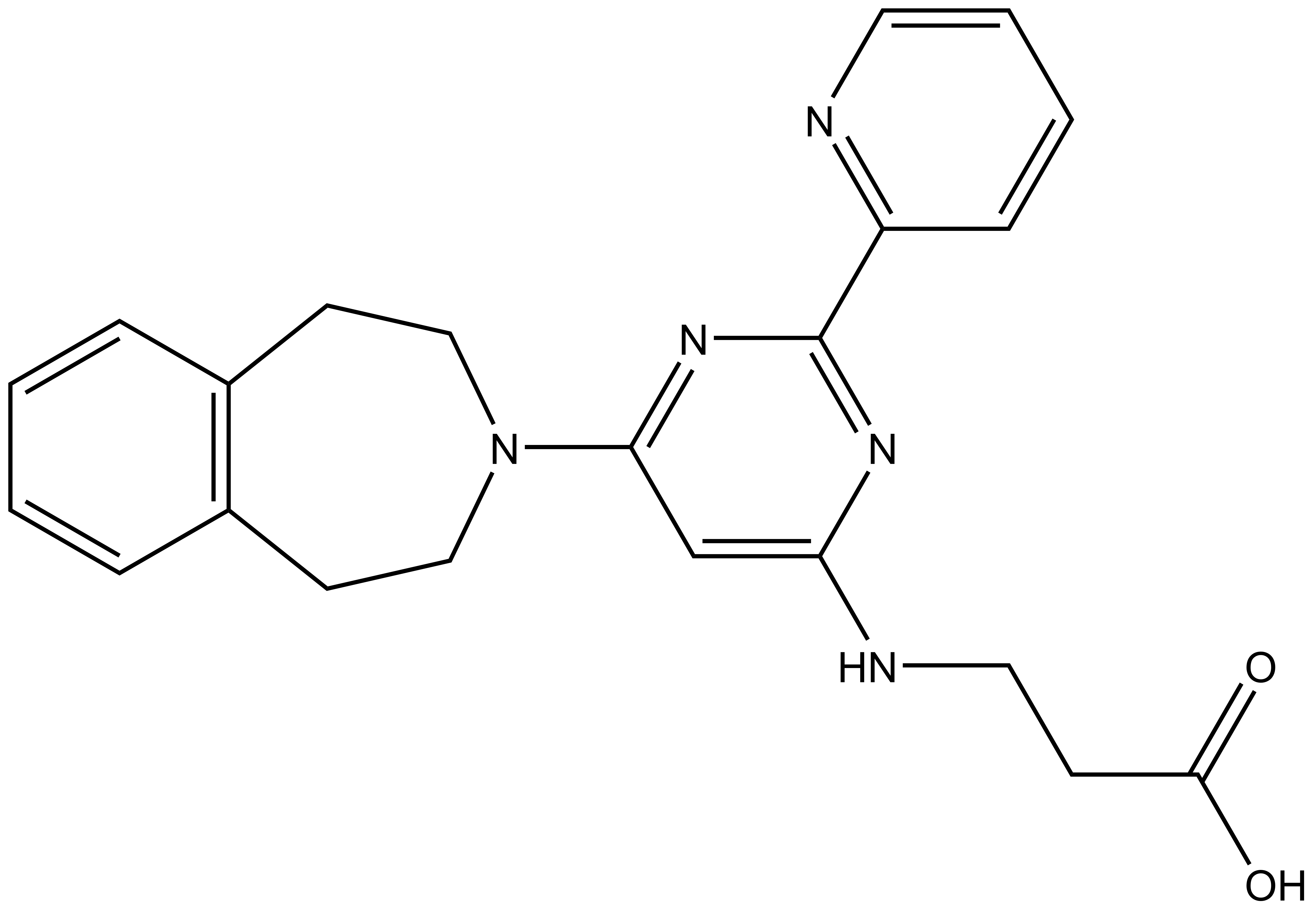 |
| 3-{[2-(pyridin-2-yl)-6-(2,3,4,5-tetrahydro-1H-3-benzazepin-3-yl)pyrimidin-4-yl]amino}propanoic acid Click here to download SDF file |
| Physical and chemical properties | |
|---|---|
| Molecular weight | 389.46 |
| Molecular formula | C22H23N5O2 |
| IUPAC name | 3-{[2-(pyridin-2-yl)-6-(2,3,4,5-tetrahydro-1H-3-benzazepin-3-yl)pyrimidin-4-yl]amino}propanoic acid |
| clogP | 2.23 |
| PSA | 91.24Å2 |
Compounds GSK-J1 and GSK-J2 were screened against a range of Jmj enzymes using the specified assays. GSK-J1 also does not significantly inhibit 100 protein kinases at a concentration of 30 µM in a competition binding assay and had negligible off-target activity against a panel of 60 unrelated proteins, including other chromatin-modifying enzymes such as histone deacetylases (see also Supplementary Table 4 and 5, data taken from primary reference (Kruidenier et al., Nature 2012)).
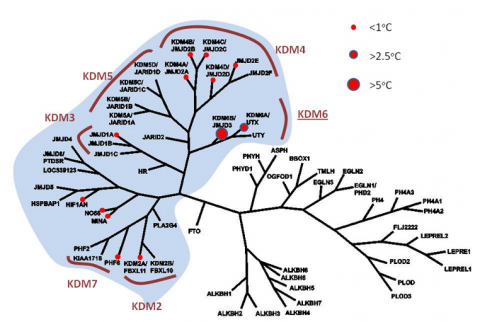
Profiling of GSK-J1 in a Tm shift assay (differential scanning fluorimetry) against a panel of human 2’OG oxygenases (see phylogenetic profile) reveals significant Tm shifts with H3K27me3 demethylases (> 2.5 C) but not with any other member, suggesting no significant crossreactivities for this H3K27 demethylase inhibitor. Known histone lysine demethylase subfamilies (KDM) are indicated. Recent alphascreen data has shown that GSK-J1 can also inhibit the H3K4me3/2/1 demethylases JARID1B/C (see table).
| Selectivity | ||||||
|---|---|---|---|---|---|---|
| Alpha Screen | GSK-J1 Mean IC50 (µM) | GSK-J2 Mean IC50 (µM) | 2-OG concentration (µM) | Fe2+ Final Concentration (µM) | Peptide substrate and concentration (µM) | AlphaScreen Detection Antibody |
| Human JmjD3 | 0.06 | >100 | 10 | 10 | Biotin-H3(14-34)K27Me3 0.06 µM | Millipore 07-452 (0.4 mg/ml) |
| Human JmjD2a | 44.3 | - | 10 | 1 | Biotin-H3(1-15)K9Me3 0.03 µM | Abcam ab1220 (0.05 mg/ml) |
| Human JmjD2e | 19.6 | - | 10 | 1 | Biotin-H3(1-15)K9Me3 0.03 µM | Abcam ab1220 (0.05 mg/ml) |
| Human JmjD1a | 41.0 | - | 5 | 10 | H3(1-21)K9Me2-Biotin 0.06 µM | Abcam ab8896 (0.4 mg/ml) |
| Jarid1b | 0.95 | 5 | 10 | H3(1-21)K4Me3-Biotin 0.1 µM | Cell Signaling Technology 9726S (1:3000) | |
| Jarid1c | 1.76 | 5 | 10 | H3(1-21)K4Me3-Biotin 0.1 µM | Cell Signaling Technology 9726S (1:3000) | |
Histone Demethylase AlphaScreen. Inhibition of histone demethylases was assessed using the histone demethylase AlphaScreen assay (Amplified Luminescence Proximity Homogenous Assay) (Kawamura et al. 2010). The demethylase AlphaScreen assays were performed in 384-well plate format using white proxiplates (Perkin Elmer). All reagents were from Sigma Aldrich unless otherwise stated and were of the highest purity. Bovine serum albumin (BSA) used in the AlphaScreen assays was fatty acid and globulin free (Sigma A7030). Hydroxy Ethyl Piperazine Ethane Sulfonic acid (HEPES) buffer was from Apollo Scientific. All steps were carried out in assay buffer (50 mM HEPES pH 7.5, 0.1% (w/v) BSA and 0.01 % (v/v) Tween- 20). FAS was dissolved fresh each day in 20 mM HCl to a concentration of 400 mM and diluted to 1.0 mM in deionized water. All other components were dissolved fresh each day in deionized water. For IC50 determinations 5 µl of assay buffer containing demethylase enzyme was transferred to wells of a 384-well proxiplate. Final assay concentrations of enzymes were: JMJD3 (1 nM), JMJD2E (2 nM), JMJD2C (5 nM), JMJD1A (0.2 nM), JARID1B (0.5 nM), JARID1C (0.5 nM). Titrations of compound (0.1 µl) were transferred to each well and the enzymes pre-incubated for 15 minutes with compound (final concentration of DMSO was 1%). The enzyme reaction was initiated by addition of 5 µl of a substrate mix consisting of aKG (see table for final assay concentration), FAS (see table for final assay concentration), L-Ascorbic Acid (100 µM) and biotinylated peptide substrate (see table for details and final assay concentrations). Enzyme reactions were allowed to proceed for the following times: JMJD3 (5 min), JMJD2E (20 min), JMJD2C (25 min), JMJD1A (5 min), JARID1B (15 min), JARID1C (15 min). The enzyme reaction was stopped after the indicated time by addition of 5 ml of EDTA (7.5 mM final assay concentration). Streptavidin Donor beads (0.08 mg/ml) and Protein-A conjugated acceptor beads (0.08 µg/ml) were pre-incubated for 1 hour with an antibody to the product methyl mark (see table for details) and the presence of the product methyl mark was detected by addition of 5 ml of the pre-incubated alphascreen beads (final concentrations of 0.02 mg/ml with respect to acceptor and donor beads). Detection was allowed to proceed for 1 hour at room temperature and the assay plates read in a BMG Labtech Pherastar FS plate reader. Data were normalized to the no enzyme control and the IC50 determined from the nonlinear regression curve fit using GraphPad Prism 5.
Profiling across Fe2+ / 2-Oxoglutarate Oxygenases. The binding of inhibitors was profiled across a range of Fe2+ / KG dependent oxygenases by using melting temperature (Tm) shift analysis (Niesen et al. 2007) to assess changes in the thermal stability upon binding of inhibitor. Protein unfolding was monitored in the presence of the fluorophore SYPRO Orange (Invitrogen) that has a strong fluorescent signal when bound to hydrophobic patches as a protein unfolds. The Tm shift assay was performed in 96-well plate format using white thin walled 0.2 ml PCR plates (Appleton Woods). Sypro Orange dye (Invitrogen) was diluted 1:1000 in 50 µM HEPES pH7.4, 150 µM NaCl, 50 µM NiCl2 and enzyme added to a concentration of 1 µM. For Tm shift assays 19 µl of enzyme was transferred to each well of a 96-well PCR plate and compound (1.0 µl) added to a final concentration of 20 µM (final DMSO concentration was 0.5%). Thermal unfolding of protein was monitored over 71 cycles in a Stratagene Mx3005p real time PCR machine by increasing the temperature by 3°C every minute. Increase in fluorescence was monitored at 492 nM excitation and 610 nM emission. Fluorescence intensity values were plotted as a function of temperature in GraphPad Prism 5 and Tm values were calculated by fitting the data to the Boltzmann equation.
For further experimental details, please see the supplementary information.
The effects of GSK-J4 (ethyl ester derivative of GSK-J1) were assessed in LPS stimulated human macrophages derived from monocytes or PBMC- in this system the compound shows an inhibitory effect on pro-inflammatory cytokine (e.g. TNF production) with an IC 50 of 9 µM. No significant adverse effects were observed at 20 µM in proliferating human primary fibroblasts.
| Assay | System | IC50 |
|---|---|---|
| Inhibition of TNF release | Primary human macrophages derived from PBMC (healthy volunteers or rheumatoid arthritis patients) | 9 µM |
| Increase in global H3K27me3 levels (immunofluorescence) | HeLa cells | < 10 µM (estimated) |
A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Laurens Kruidenier, Chun-wa Chung, Zhongjun Cheng, John Liddle, KaHing Che, Gerard Joberty, Marcus Bantscheff, Chas Bountra, Angela Bridges, Hawa Diallo, Dirk Eberhard, Sue Hutchinson, Emma Jones, Roy Katso, Melanie Leveridge, Palwinder K. Mander, Julie Mosley, Cesar Ramirez-Molina, Paul Rowland, Christopher J. Schofield, Robert J. Sheppard, Julia E. Smith, Catherine Swales, Robert Tanner, Pamela Thomas, Anthony Tumber, Gerard Drewes, Udo Oppermann, Dinshaw J. Patel, Kevin Lee & David M. Wilson.
Nature, 2012, 488, 404–408