The probe is available from Sigma and Cayman Chemicals.
The control may be requested here.
| Probe | Negative control | Negative control | ||
 |
|  |
|  |
Homer |
| nc_WDR5 |
| nc_VHL |
The Histone Lysine Methyltransferase (HMT) complex MLL1 is a crucial epigenetic writer to control DNA accessibility and promote gene transcription [1,2]. The complex incorporates WDR5 (WD40-repeat containing protein 5) that acts as scaffolding component and enhances HMT activity of MLL1. [2] The propeller-shaped WDR5 protein contains two binding sites that interact not only with oncogenic drivers like MLL1, but also with various other proteins like the oncoprotein MYC. [3]
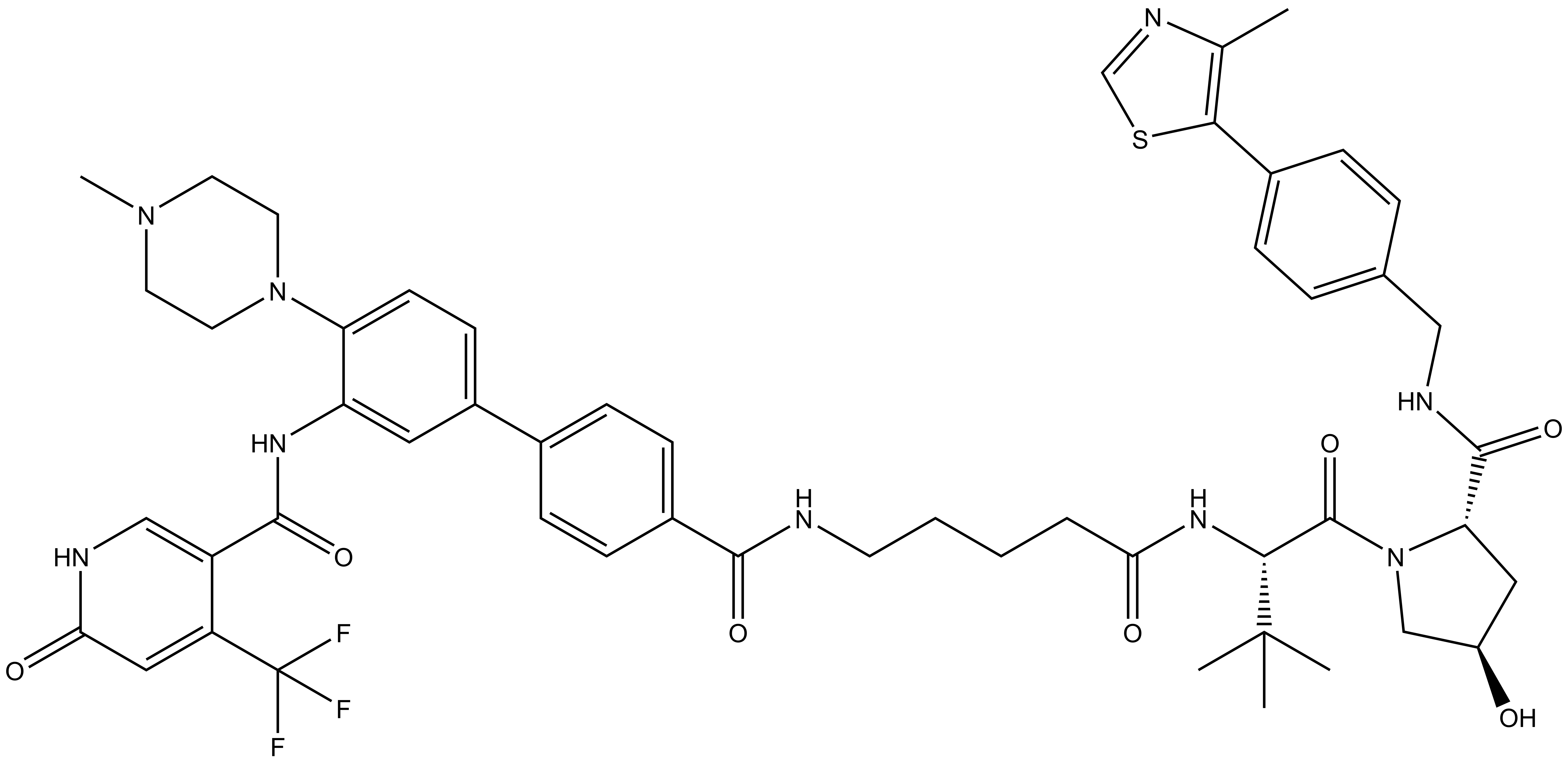
Here, we present a new chemical degrader probe for the WDR5 protein, Homer, that was initially designed by the OICR and the SGC Toronto [4, 5] and then further advanced into a PROTAC (Proteolysis targeting chimera) chemical probe by the SGC Frankfurt [6]. This scaffold represents the first chemical degrader probe for WDR5.
Homer binds potently to WDR5 with a K D (ITC) of 18 nM. It is selective as shown in quantitative proteomic studies. This compound is non-toxic as demonstrated by the AlarmarBlue assay in U2OS cells. Homer shows remarkably cellular activity in MV4-11 cells by inducing WDR5 degradation in a catalytic manner a DC 50 of 53 nM. The DC max of Homer is 58% and observed at cellular concentrations of 1 µM.
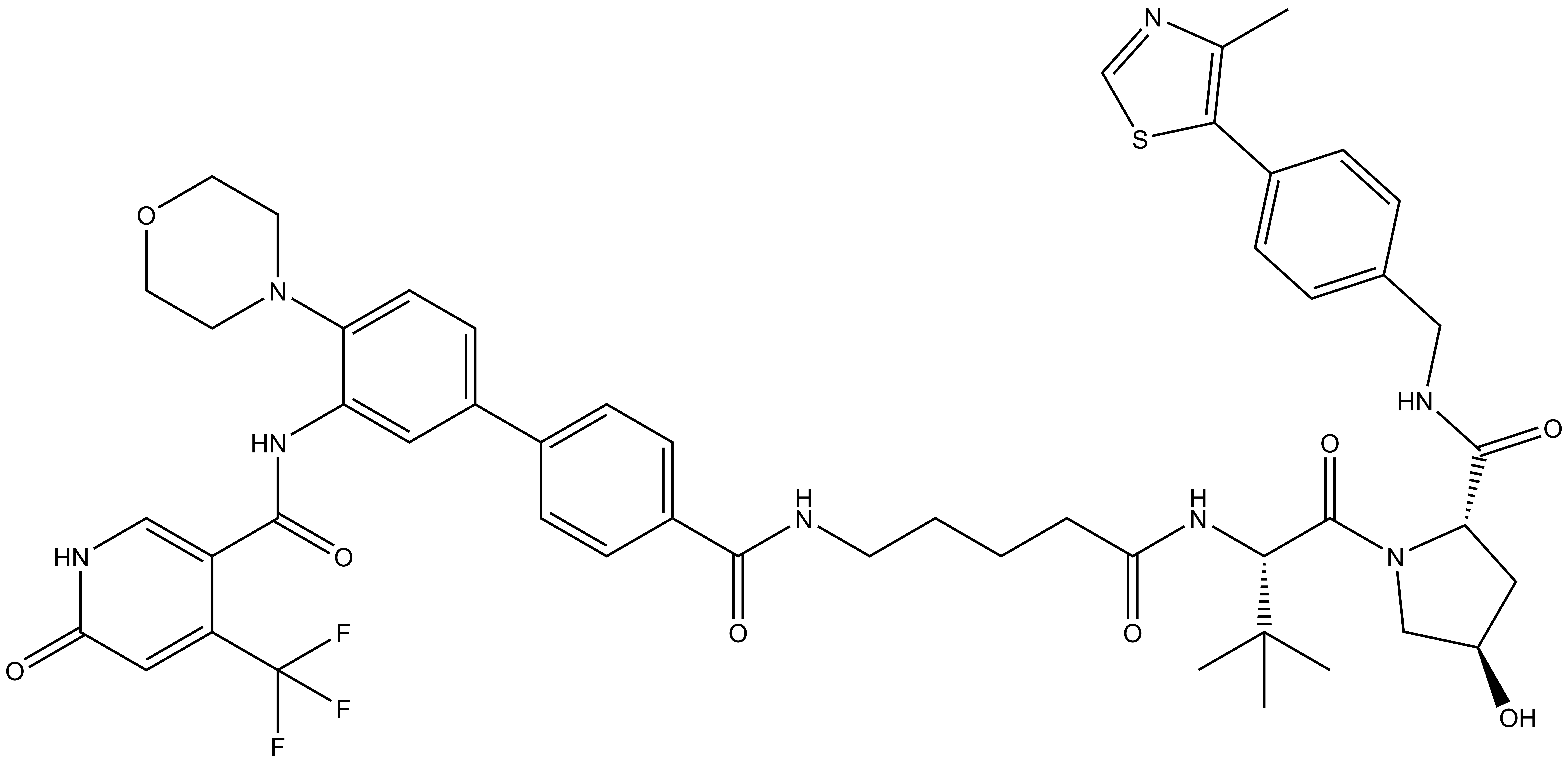
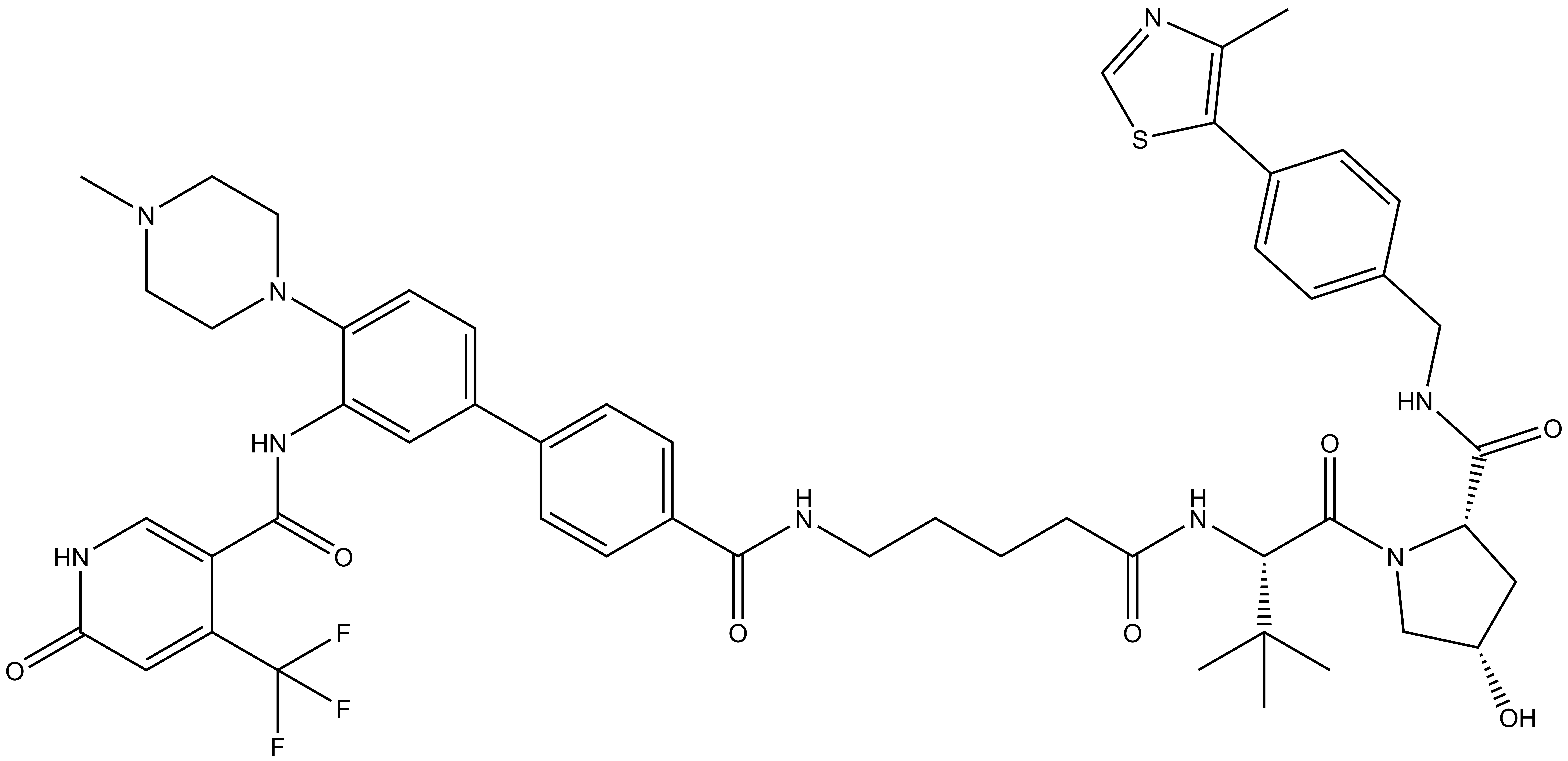
In addition, two negative control compounds, nc_WDR5 and nc_VHL, are provided, which showed no degradational activity.
Summary of Potency and efficacy of Homer
| WDR5 | Homer KD |
| DSF | 13.2 K |
| ITC | 18 nM |
| NanoBRET | 13.6 µM |
| HiBit | DCmax: 58% at 1 µM DC50: 53 nM |
| quantitative Proteomics | (-log10p > 3, log2FC < -0.5) |
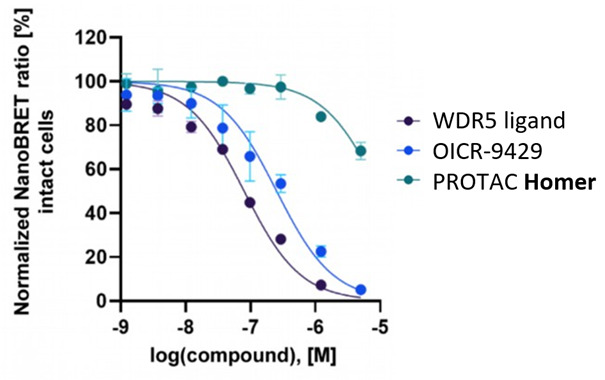
Selectivity of the first WDR5 probe OICR-9429 was shown previously [5, 6]. Selectivity for the Homer probe was confirmed by quantitative proteomics.
For optimal degradation and to overcome the Hook effect, we recommend that a concentration of 1 µM should be used in cell-based assays.
Homer induces depletion of endogenous WDR5 in various cancer cell lines by inducing protein ubiquitylation and degradation.
| Probe | Negative control | Negative control | ||
 |
|  |
|  |
Homer |
| nc_WDR5 |
| nc_VHL |
| Homer | |
| Physical and Chemical Properties | |
| Molecular weight | 1012.16 |
| Molecular formula | C52H60F3N9O7S |
| IUPAC name | N-(4'-((5-(((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-5-oxopentyl)carbamoyl)-4-(4-methylpiperazin-1-yl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide |
| clogPo/w (SwissADME) | 5.31 |
| TPSA (SwissADME) | 237.41 Å2 |
| No. of chiral centres | 3 |
| No. of rotatable bonds | 23 |
| No. of hydrogen bond acceptors | 12 |
| No. of hydrogen bond donors | 6 |
| Storage | stable as powder at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | soluble in DMSO at 50 mM |
| nc_VHL | |
| Physical and Chemical Properties | |
| Molecular weight | 1012.16 |
| Molecular formula | C52H60F3N9O7S |
| IUPAC name | N-(4'-((5-(((S)-1-((2S,4S)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-5-oxopentyl)carbamoyl)-4-(4-methylpiperazin-1-yl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide |
| clogPo/w (SwissADME) | 5.25 |
| TPSA (SwissADME) | 237.41 Å2 |
| No. of chiral centres | 3 |
| No. of rotatable bonds | 23 |
| No. of hydrogen bond acceptors | 12 |
| No. of hydrogen bond donors | 6 |
| Storage | stable as powder at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | soluble in DMSO at 50 mM |
| nc_WDR5 | |
| Physical and Chemical Properties | |
| Molecular weight | 999.12 |
| Molecular formula | C51H57F3N8O9S |
| IUPAC name | N-(4'-((5-(((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-5-oxopentyl)carbamoyl)-4-morpholino-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide |
| clogPo/w (SwissADME) | 5.41 |
| TPSA (SwissADME) | 243.40 Å2 |
| No. of chiral centres | 3 |
| No. of rotatable bonds | 23 |
| No. of hydrogen bond acceptors | 12 |
| No. of hydrogen bond donors | 6 |
| Storage | stable as powder at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | soluble in DMSO at 50 mM |
Homer: CC1=C(SC=N1)C2=CC=C(C=C2)CNC([C@@H]3C[C@H](CN3C([C@@H](NC(CCCCNC(C4=CC=C(C=C4)C5=CC=C(C(NC(C6=CNC(C=C6C(F)(F)F)=O)=O)=C5)N7CCN(CC7)C)=O)=O)C(C)(C)C)=O)O)=O
nc_VHL: CC1=C(C2=CC=C(CNC([C@@H]3C[C@H](O)CN3C([C@H](C(C)(C)C)NC(CCCCNC(C4=CC=C(C5=CC=C(N6CCN(C)CC6)C(NC(C7=CNC(C=C7C(F)(F)F)=O)=O)=C5)C=C4)=O)=O)=O)=O)C=C2)SC=N1
nc_WDR5: CC1=C(C2=CC=C(CNC([C@@H]3C[C@@H](O)CN3C([C@H](C(C)(C)C)NC(CCCCNC(C4=CC=C(C5=CC=C(N6CCOCC6)C(NC(C7=CNC(C=C7C(F)(F)F)=O)=O)=C5)C=C4)=O)=O)=O)=O)C=C2)SC=N1
Homer: InChI=1S/C52H60F3N9O7S/c1-31-45(72-30-59-31)34-11-9-32(10-12-34)27-58-49(70)42-25-37(65)29-64(42)50(71)46(51(2,3)4)61-43(66)8-6-7-19-56-47(68)35-15-13-33(14-16-35)36-17-18-41(63-22-20-62(5)21-23-63)40(24-36)60-48(69)38-28-57-44(67)26-39(38)52(53,54)55/h9-18,24,26,28,30,37,42,46,65H,6-8,19-23,25,27,29H2,1-5H3,(H,56,68)(H,57,67)(H,58,70)(H,60,69)(H,61,66)/t37-,42+,46-/m1/s1
nc_VHL: InChI=1S/C52H60F3N9O7S/c1-31-45(72-30-59-31)34-11-9-32(10-12-34)27-58-49(70)42-25-37(65)29-64(42)50(71)46(51(2,3)4)61-43(66)8-6-7-19-56-47(68)35-15-13-33(14-16-35)36-17-18-41(63-22-20-62(5)21-23-63)40(24-36)60-48(69)38-28-57-44(67)26-39(38)52(53,54)55/h9-18,24,26,28,30,37,42,46,65H,6-8,19-23,25,27,29H2,1-5H3,(H,56,68)(H,57,67)(H,58,70)(H,60,69)(H,61,66)/t37-,42-,46+/m0/s1
nc_WDR5: InChI=1S/C51H57F3N8O8S/c1-30-44(71-29-58-30)33-10-8-31(9-11-33)26-57-48(68)41-24-36(63)28-62(41)49(69)45(50(2,3)4)60-42(64)7-5-6-18-55-46(66)34-14-12-32(13-15-34)35-16-17-40(61-19-21-70-22-20-61)39(23-35)59-47(67)37-27-56-43(65)25-38(37)51(52,53)54/h8-17,23,25,27,29,36,41,45,63H,5-7,18-22,24,26,28H2,1-4H3,(H,55,66)(H,56,65)(H,57,68)(H,59,67)(H,60,64)/t36-,41+,45-/m1/s1
Homer: OFNZESNEBSQKSE-BQGOKDIQSA-N
nc_VHL: OFNZESNEBSQKSE-CCVFZADKSA-N
nc_WDR5: BFXBMIZHEIFXOC-LTJJNQLXSA-N
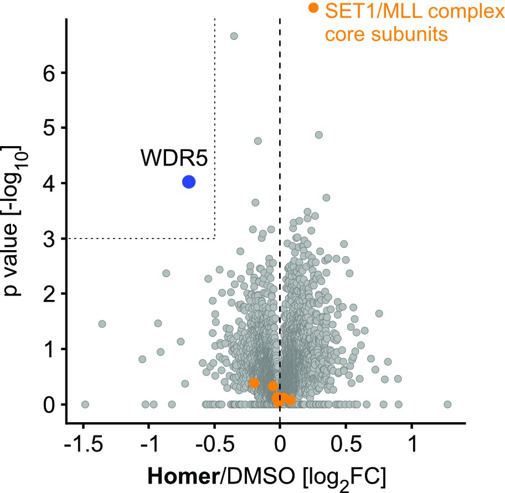 |
| Quantitative Proteomics revealed that only WDR5 is significally and substantially depleted by Homer treatment. |
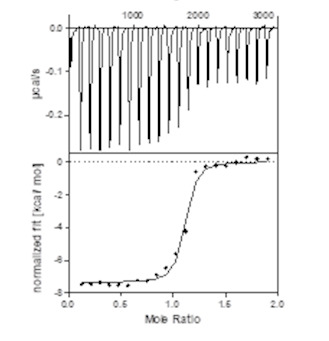 |
| Homer binds potently to WDR5 as determined by ITC. |
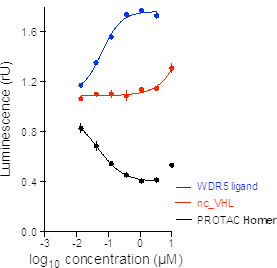
Homer is cell permeable and degrades WDR5 in MV4-11 cells. The control compounds nc_WDR5 and nc_VHL are not active.
NanoBRET data of Homer in HEX293 cells show Homer is cell permeable.
 | 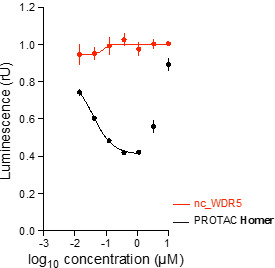 |
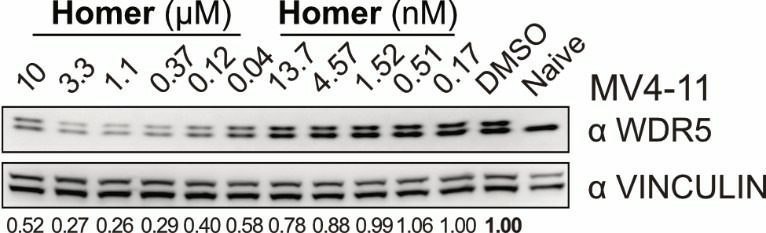 | 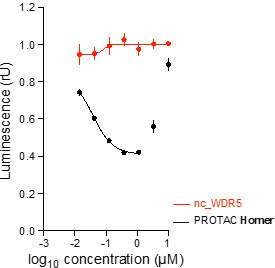 |
| (up) HiBiT assay curves from Homer (black) and both negative controls (red) as well as from parent WDR5 compound (blue) show that Homer degrades WDR5 in MV4-11 cells after 24 h. The most effective degradation (58% WDR5 depletion) is observed at 1 µM. (down) WesternBlots validate HiBiT data and show degradation of WDR5 by Homer treatment in MV4-11 cells after 24 h. | |
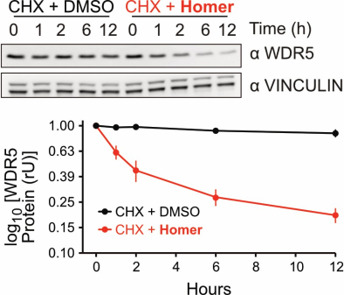
Homer decreases WDR5 protein stability as shown in the Cycloheximide chase assay.
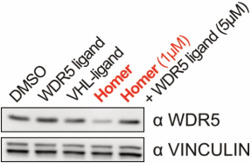
Rescue experiments show that WDR5 depletion requires binding of Homer. WDR5 levels can be restored by excess of WDR5 ligand.
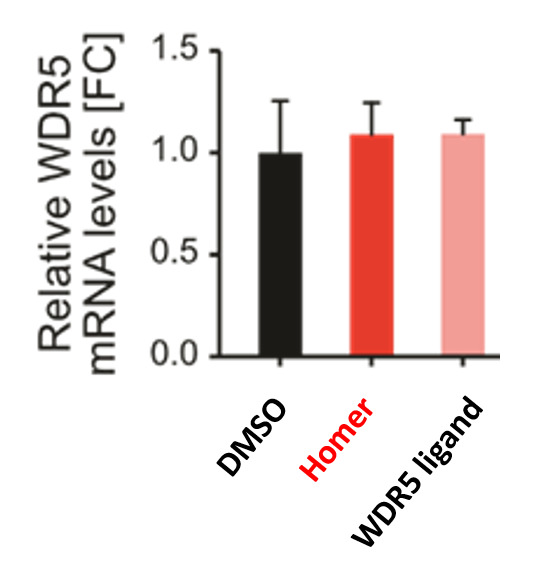
qPCR shows that Homer does not affect WDR5 transcription.
[1] Wu, Shu, “MLL1/WDR5 complex in leukemogenesis and epigenetic regulation”, Chin. J. Cancer 2011, 30(4), 240-246.
[2] Jiang, “The complex activities of the SET1/ MLL complex core subunits in development and disease“, BBA – Gene Regulatory Mechanisms 2020, 1863, 194560.
[3] Thomas et al., “Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC”, Mol. Cell 2015, 58, 440-452.
[4] Grebien et al., “Pharmacological targeting of the Wdr5-MLL interaction in C/EBPa N-terminal leukemia”, Nat Chem Biol. 2015, 11, 571.
[5] Getlik et al., “Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1)”, J Med Chem. 2016, 59, 2478.
[6] Dölle, Adhikari et al., “Design, Synthesis, and Evaluation of WD-Repeat-Containing Protein 5 (WDR5) Degraders”, J Med Chem. 2021, May 13. doi: 10.1021/acs.jmedchem.1c00146. Epub ahead of print. PMID: 33980013.
[7] A.C. Wallace, R.A. Laskowski, J.M. Thornton, “LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions”, Protein Eng. 8, 1995, 127-134.
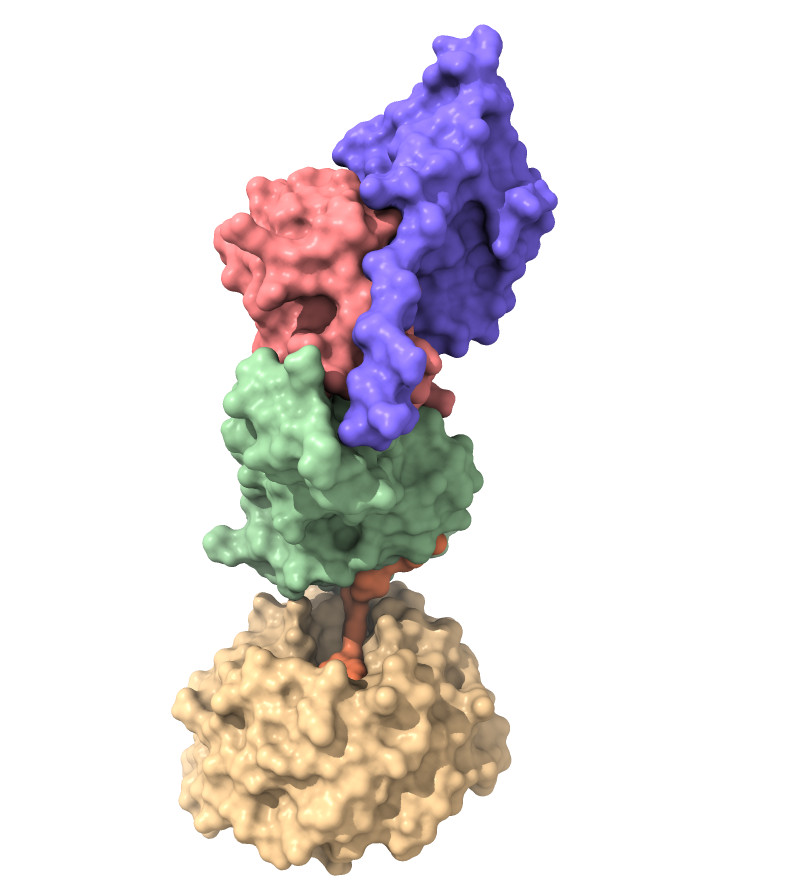 | 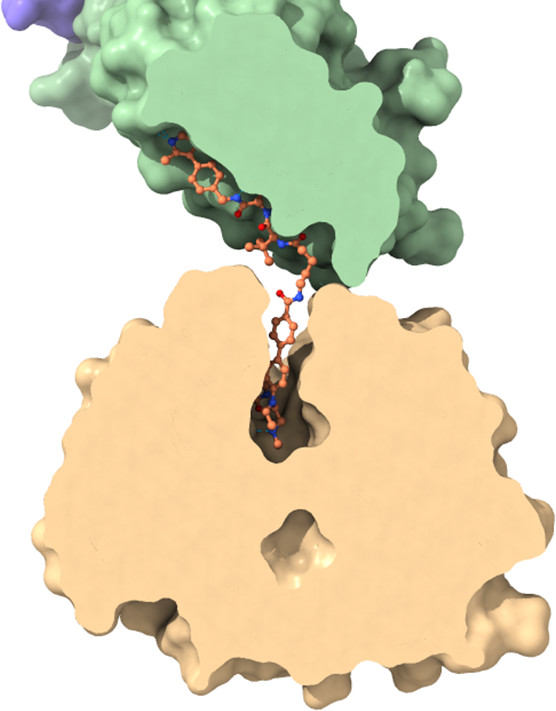 |
Left panel: Quaternary complex of WDR5:VHL:ElonginB:ElonginC:Homer bound to chemical probe PROTAC Homer in surface representation.
Right panel: Surface slice with focus on the binding pocket of WDR5 and VHL. Homer is shown in stick representation
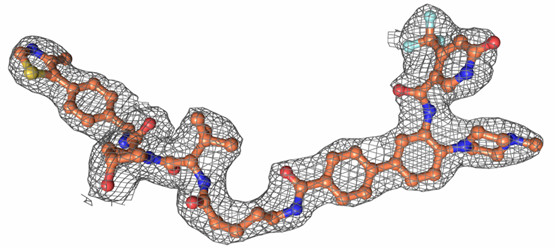
Observed electron density of Homer countered at 1 sigma.
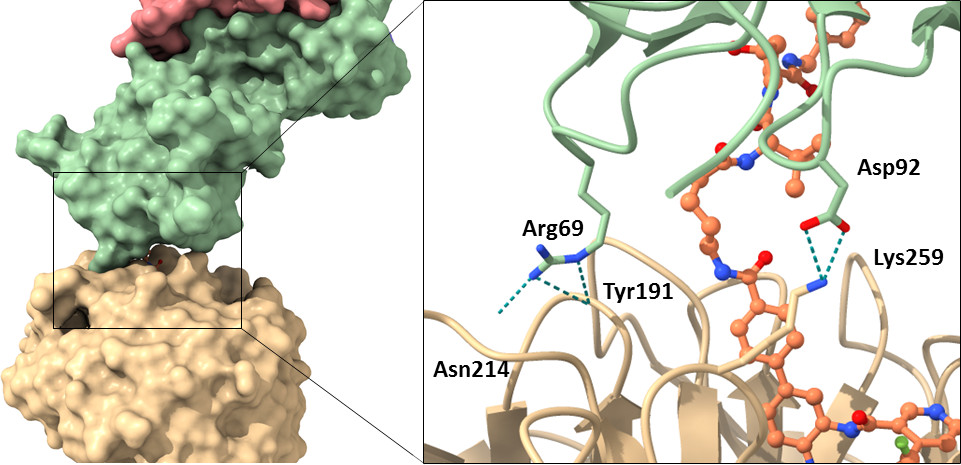
Left panel: Contact surface area between WDR5 (wheat) and VHL (green) in surface representation
Right panel: Close up on the detailed interaction between WDR5 and VHL in cartoon/stick representation. The orientation is the same as in the left Panel.
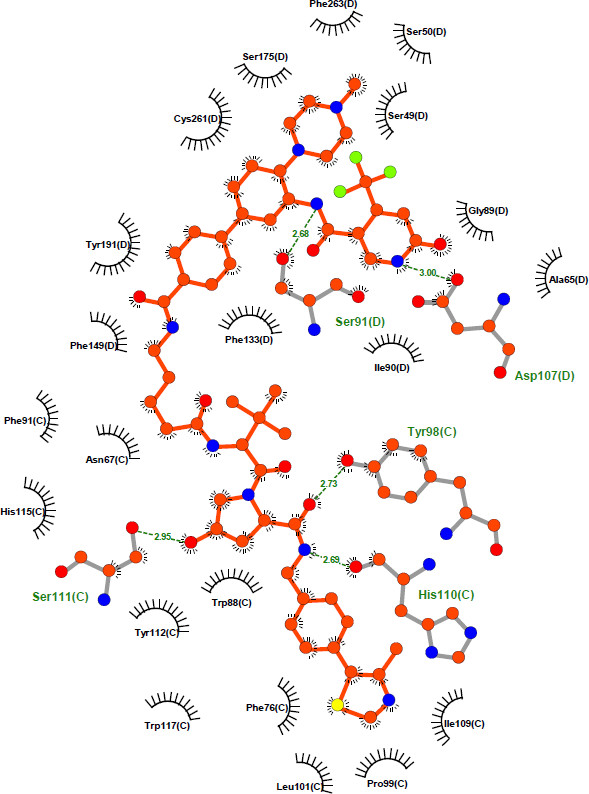 | WDR5 (D) |
| VHL (C) |
Detailed interaction between Homer and VHL:WDR5. The figure was created with LIGPLOT [7]