| Probe | Negative control | |
 |
|  |
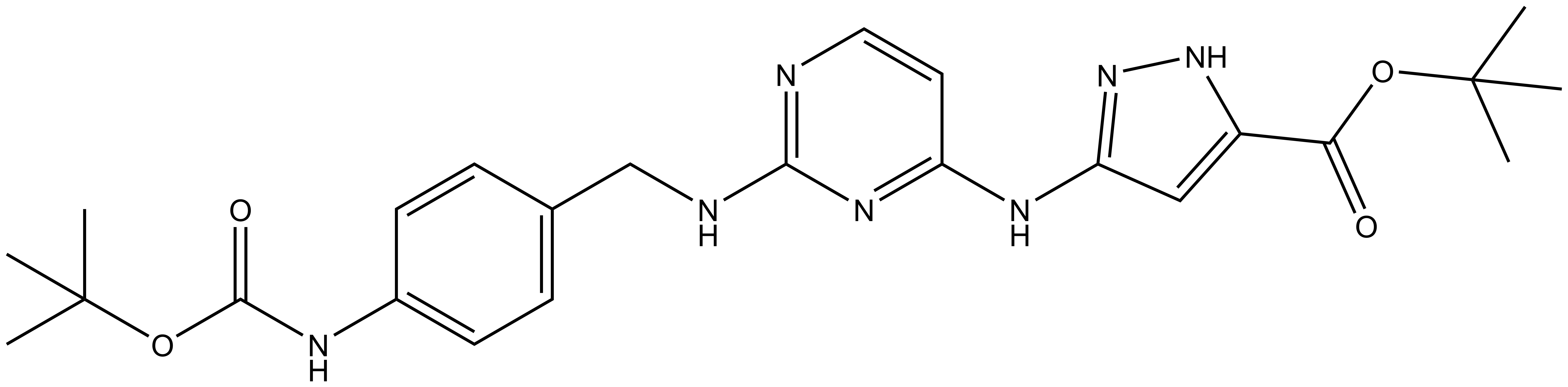
JA397 |
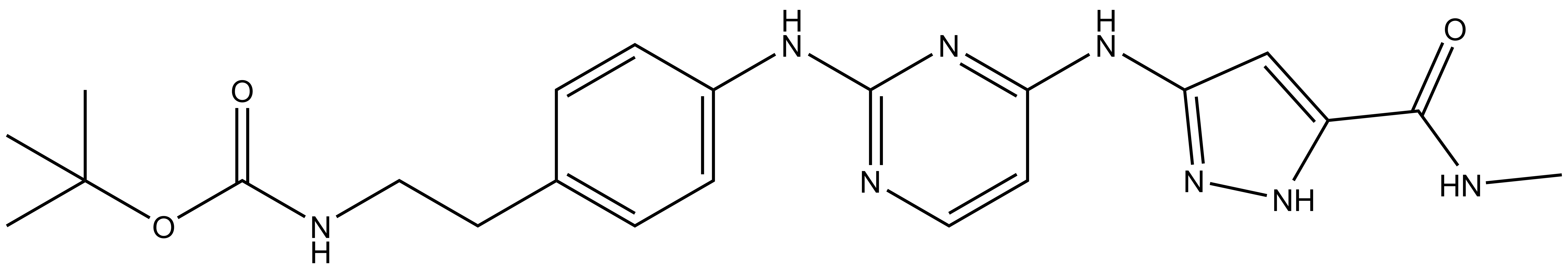
| JA314 |
The TAIRE family of protein kinases are the cyclin-dependent kinases (CDK14-18), which are Ser/Thr kinases and belong to the CMGC family. The CDKs are activated by specific cyclins and are known to play an important role in cell cycle regulation (Wood 2018). Therefore, CDKs are now an important protein family that also has therapeutic significance, as demonstrated by the recent FDA-approved drugs for CDK4/6. However, for the TAIRE family little is known. However, the TAIRE family is less well described in the literature. It can be further subdivided into the PFTAIRE family (CDK14-15) and PCTAIRE family (CDK16-18). There is various evidence, for example, that CDK14 is involved in the WNT signalling pathway (Davidson, 2010); CDK15 regulates the beta-catenin/MEK-ERK signalling pathway (Huang, 2015), among others; CDK16 has an effect on cell cycle through phosphorylation of P27 (Yanagi, 2016); CDK17 plays a role in glycerophospholipid metabolism (Liu, 2017); and CDK18 regulates cell motility through the FAK/RhoA/ROCK signalling pathway (Matsuda, 2017).
The SGC has developed JA397, a potent and selective inhibitor for the TAIRE family with cellular activity, ranging from IC50 values of 21 nM to 307 nM as determined by NanoBRET.
The chemical probe (JA397) is accompanied by a negative control (JA314) that is structurally closely related to the probe molecule.
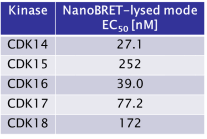 |
JA397 had an EC50 of 27.1 nM, 252 nM, 39.0 nM, 77.2 nM and 172 nM to CDK14, CDK15, CDK16, CDK17 and CDK18, respectively in the NanoBRET-lysed mode assay.
JA397 was selective in an in vitro kinase panel from Reaction Biology at 1 µM against 340 WT Kinases, followed by cellular NanoBRET assays. Selectivity within the CDK family was determined by NanoBRET.
Based on the potency and the selectivity of the chemical probe and to minimize the risk of unspecific cytotoxicity, we recommend a concentration of no higher than 1 µM for cell-based assays.
JA397 displayed an EC50 of 72.1 nM, 307 nM, 33.4 nM, 21.2 nM and 121 nM on CDK14, CDK15, CDK16, CDK 17 and CDK18 respectively in intact cells in the NanoBRET assay.
| Probe | Negative control | |
 |
|  |
JA397 |
| JA314 |
| Physical and chemical properties JA397 | |
| Molecular weight | 481.56 |
| Molecular formula | C24H31N7O4 |
| IUPAC name | tert-butyl 3-((2-((4-((tert-butoxycarbonyl)amino)benzyl)amino)pyrimidin-4-yl)amino)-1H-pyrazole-5-carboxylate |
| clogP | 4.86 |
| tPSA | 137.8 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 11 |
| No. of hydrogen bond acceptors | 10 |
| No. of hydrogen bond donors | 4 |
| Storage | r. t. |
SMILES: O=C(C1=CC(NC2=NC(NCC3=CC=C(C=C3)NC(OC(C)(C)C)=O)=NC=C2)=NN1)OC(C)(C)C
InChI: InChI=1S/C24H31N7O4/c1-23(2,3)34-20(32)17-13-19(31-30-17)28-18-11-12-25-21(29-18)26-14-15-7-9-16(10-8-15)27-22(33)35-24(4,5)6/h7-13H,14H2,1-6H3,(H,27,33)(H3,25,26,28,29,30,31)
InChIKey: JQLMEZBHIJSVKR-UHFFFAOYSA-N
| Physical and chemical properties JA314 | |
| Molecular weight | 452,52 |
| Molecular formula | C22H28N8O3 |
| IUPAC name | tert-butyl (4-((4-((5-(methylcarbamoyl)-1H-pyrazol-3-yl)amino)pyrimidin-2-yl)amino)phenethyl)carbamate |
| clogP | 3.55 |
| tPSA | 140.6 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 10 |
| No. of hydrogen bond acceptors | 9 |
| No. of hydrogen bond donors | 5 |
| Storage | r. t. |
SMILES: O=C(C1=CC(NC2=NC(NC3=CC=C(C=C3)CCNC(OC(C)(C)C)=O)=NC=C2)=NN1)NC
InChI: InChI=1S/C22H28N8O3/c1-22(2,3)33-21(32)25-11-9-14-5-7-15(8-6-14)26-20-24-12-10-17(28-20)27-18-13-16(29-30-18)19(31)23-4/h5-8,10,12-13H,9,11H2,1-4H3,(H,23,31)(H,25,32)(H3,24,26,27,28,29,30)
InChIKey: VJWCJKXRULCPCJ-UHFFFAOYSA-N
Kinome-wide selectivity profile of JA397 was determined in our in-house kinase DSF-panel comprising 105 kinases and at Reaction Biology at 1 µM comprising 340 WT kinases.
The selectivity against a CDK family was determined by NanoBRET.
DSF-Panel against 105 kinases:
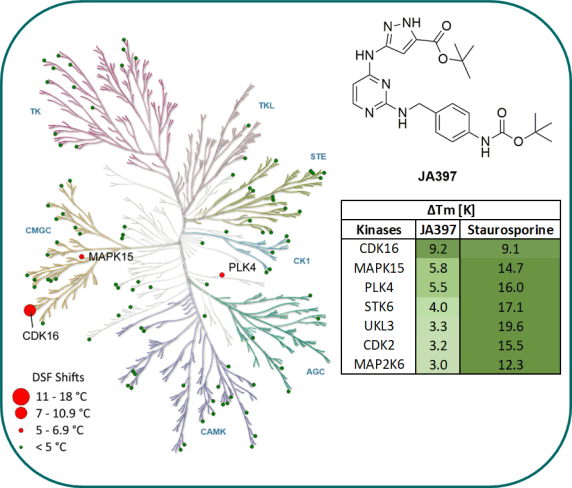 |
Reaction Biology (340 WT kinases) @ 1µM:
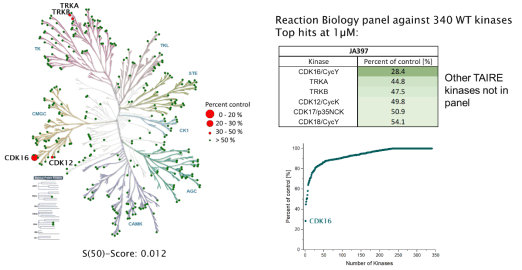 |
Selectivity within the CDK-Family:
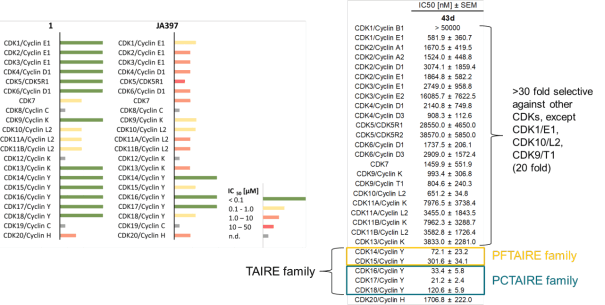 |
The negative control JA314 showed no stabilization against 105 kinases screened in our in-house DSF-Panel.
DSF-Panel against 105 kinases:
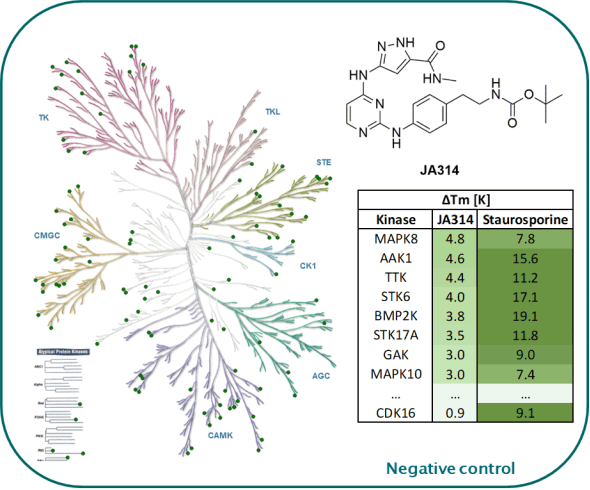 |
JA397 displayed an EC50 values ranging from 21 nM to 307 nM against the TAIRE family, determined by NanoBRETTM assay.
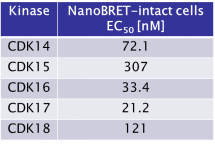 |
The negative control compound 314 displayed a EC50 value of 4165 nM against CDK16, determined by NanoBRETTM assay.
Amrhein JA, Berger LM, Tjaden A, Krämer A, Elson L, Tolvanen T, Martinez-Molina D, Kaiser A, Schubert-Zsilavecz M, Müller S, Knapp S, Hanke T. Discovery of 3-Amino-1H-pyrazole-Based Kinase Inhibitors to Illuminate the Understudied PCTAIRE Family. Int J Mol Sci. 2022 Nov 27;23(23):14834. doi: 10.3390/ijms232314834. PMID: 36499165; PMCID: PMC9736855.
Davidson G, Niehrs C. Emerging links between CDK cell cycle regulators and Wnt signaling. Trends Cell Biol. 2010 Aug;20(8):453-60. doi: 10.1016/j.tcb.2010.05.002. Epub 2010 Jun 4. PMID: 20627573.
Huang C, Du R, Jia X, Liu K, Qiao Y, Wu Q, Yao N, Yang L, Zhou L, Liu X, Xiang P, Xin M, Wang Y, Chen X, Kim DJ, Dong Z, Li X. CDK15 promotes colorectal cancer progression via phosphorylating PAK4 and regulating β-catenin/ MEK-ERK signaling pathway. Cell Death Differ. 2022 Jan;29(1):14-27. doi: 10.1038/s41418-021-00828-6. Epub 2021 Jul 14. PMID: 34262144; PMCID: PMC8738751.
Liu M, Xu Z, Du Z, Wu B, Jin T, Xu K, Xu L, Li E, Xu H. The Identification of Key Genes and Pathways in Glioma by Bioinformatics Analysis. J Immunol Res. 2017;2017:1278081. doi: 10.1155/2017/1278081. Epub 2017 Dec 6. PMID: 29362722; PMCID: PMC5736927.
Matsuda S, Kawamoto K, Miyamoto K, Tsuji A, Yuasa K. PCTK3/CDK18 regulates cell migration and adhesion by negatively modulating FAK activity. Sci Rep. 2017 Mar 31;7:45545. doi: 10.1038/srep45545. PMID: 28361970; PMCID: PMC5374530.
Wood DJ, Endicott JA. Structural insights into the functional diversity of the CDK-cyclin family. Open Biol. 2018 Sep;8(9):180112. doi: 10.1098/rsob.180112. PMID: 30185601; PMCID: PMC6170502.
Yanagi T, Matsuzawa S. PCTAIRE1/PCTK1/CDK16: a new oncotarget? Cell Cycle. 2015;14(4):463-4. doi: 10.1080/15384101.2015.1006539. PMID: 25590439; PMCID: PMC4347670.