| Probe | Negative control | |
 |
|  |
MRIA9 |
| MR7 |
Salt-inducible kinases (SIK1-3) are members of the AMP-activated protein kinase (AMPK) family which is part of the calcium/calmodulin-dependent kinase (CaMK) group. These serine/threonine kinases act as regulators of energy homeostasis and metabolic stress. The SIK family member SIK2, for example, is activated in cells recovering from starvation, leading to phosphorylation and hence activation of the transcription factor cAMP response element-binding protein (CREB1) [1-3]. In addition to the key function of SIK in regulating metabolism, imbalance of SIK has been observed in the context of several diseases, especially in cancer, with both tumor promoting and tumor suppressive roles being reported [4]. SIK2 is often deleted in breast cancer, and downregulation of SIK1 has been linked to a tumor suppressor role but also the development of metastasis by promoting p53-dependent anoikis [5]. We have recently shown that in biopsies from patients with gastric cancer, there were increased levels of SIK2 mRNA and protein in advanced stages of the tumor compared to lower grade tumors, independent of its metastatic stage [6]. Overall, these data highlight the complex roles of SIK family proteins in different types of cancer and they may emerge as important therapeutic targets. To clarify the multifaceted roles of these kinases in disease and normal physiology, chemical tools targeting SIK are urgently needed.
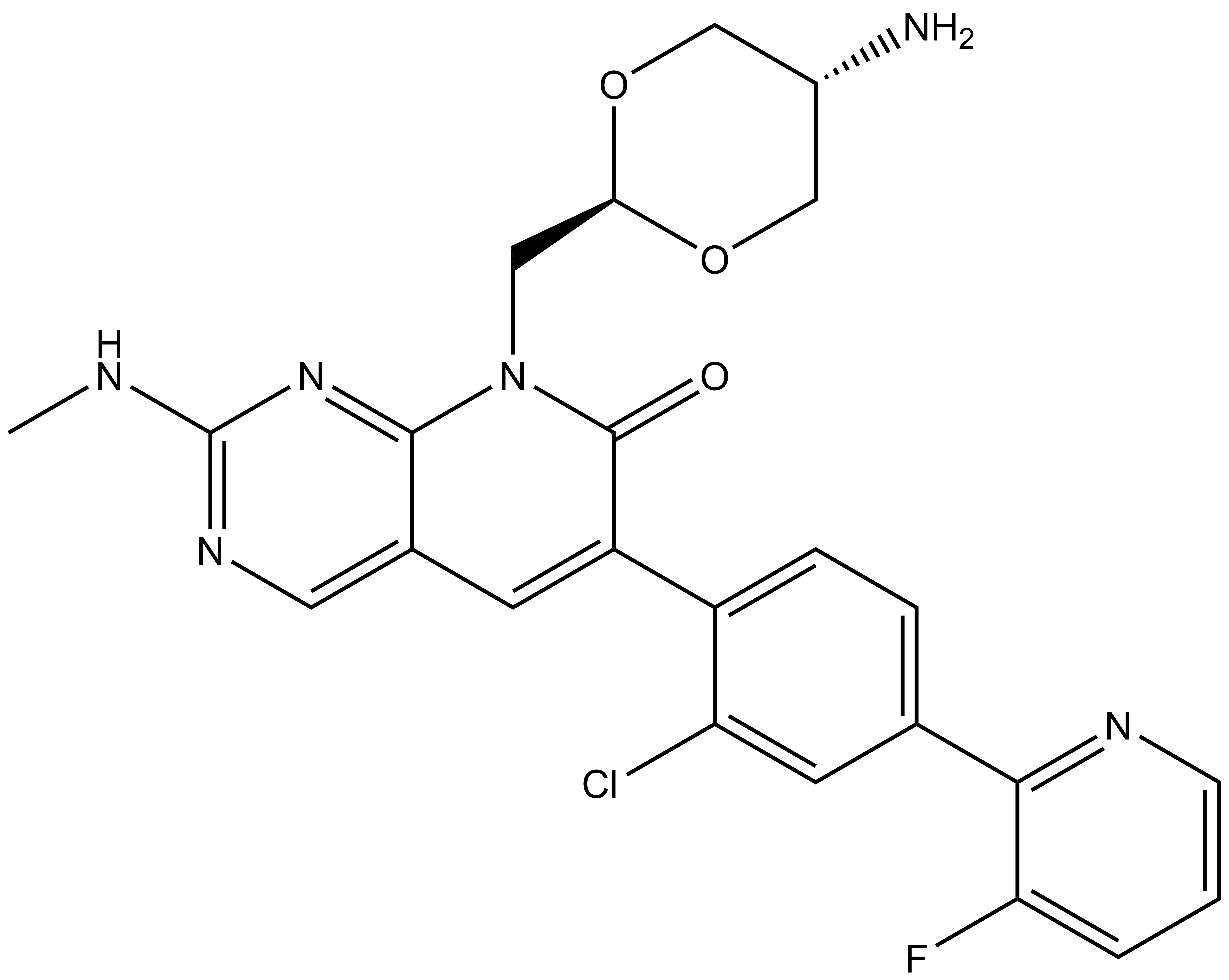
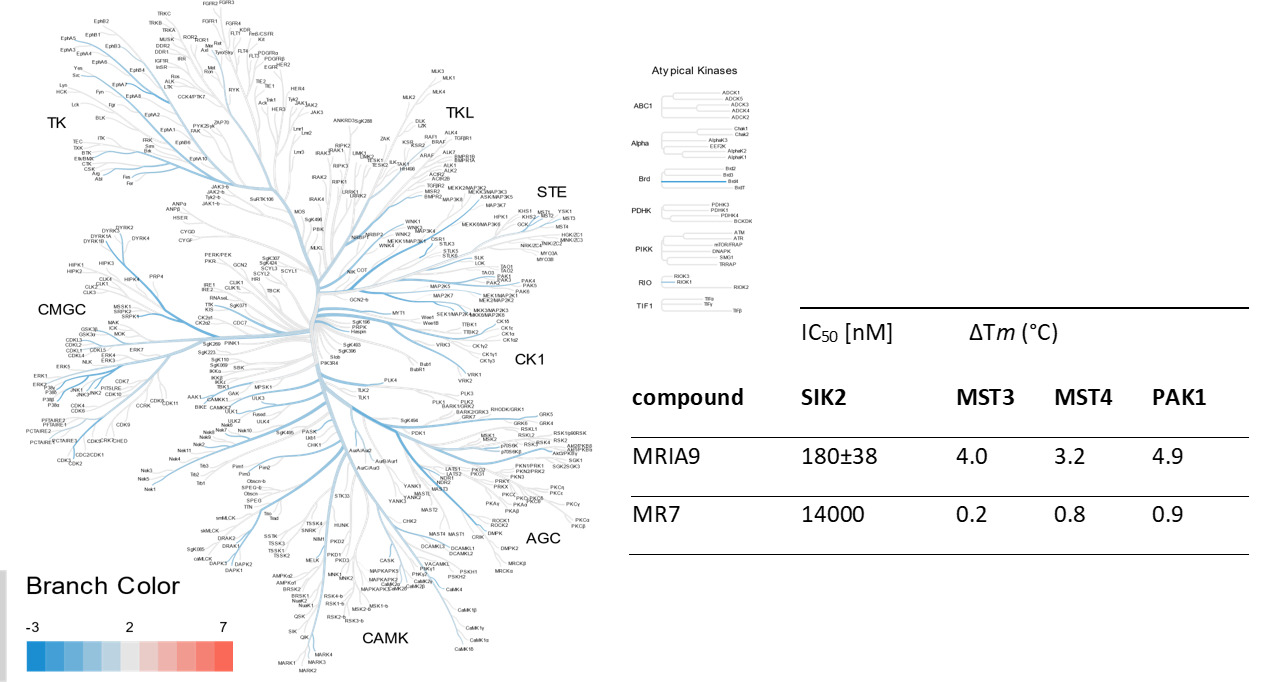
SGC has developed MRIA9, a potent and selective pan SIK inhibitor with a IC50 determined by a radiometric assay of 55, 48 and 22 nM for SIK1, SIK2 and SIK3 respectively and IC50 of 516, 180 and 127 nM on NanoBRET™ assay. [7] MRIA9 has been developed based on PAK1 inhibitor G-5555 published by Genentech. Group I PAK (PAK1, PAK2 and PAK3) remains off targets with respectively in vitro IC50s of 580, 41 and 140 nM. However, due to lack of accessibility of a NanoBRET™ assay for Group I PAKs, MRIA9 is considered a SIK and Group I PAK probe. In addition, the chemical probe (MRIA9) is accompanied by a negative control (MR7), which is structurally similar to the probe molecule.
Potency Against Target Family
| Kinase | 33PanQinase IC50 (nM) |
| SIK1 | 55 |
| SIK2 | 48 |
| SIK3 | 22 |
MRIA9 has been shown to be selective in an in vitro kinase panel from Reaction Biology followed by cellular NanoBRET assays. The selectivity outside target family revealed Group I PAKs as closest off-target.
To minimize the chance of off-target effects, we recommend a concentration of no higher than 10 µM for cell-based assays.
In NanoBRET assay using HEK923T cells MRIA9 shows an IC50 of 516, 180 and 127 nM for SIK1, SIK2 and SIK3 respectively.
| Probe | Negative control | |
 |
|  |
MRIA9 |
| MR7 |
| Physical and chemical properties MRIA9 | |
| Molecular weight | 496.93 |
| Molecular formula | C24H22ClFN6O3 |
| IUPAC name | 8-(((2r,5r)-5-amino-1,3-dioxan-2-yl)methyl)-6-(2-chloro-4-(3-fluoropyridin-2-yl)phenyl)-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one |
| logP | 2.16 |
| PSA | 113.9 |
| No. of chiral centre | 0 |
| No. of rotatable bonds | 7 |
| No. of hydrogen bond acceptors | 7 |
| No. of hydrogen bond donors | 2 |
| Storage | stable as solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | soluble in DMSO in a concentration of 50 mM |
SMILES:
CNC1=NC=C2C(N(C(C(C3=CC=C(C=C3Cl)C4=NC=CC=C4F)=C2)=O)C[C@H]5OC[C@@H](CO5)N)=N1
InChI: InChI=1S/C24H22ClFN6O3/c1-28-24-30-9-14-7-17(16-5-4-13(8-18(16)25)21-19(26)3-2-6-29-21)23(33)32(22(14)31-24)10-20-34-11-15(27)12-35-20/h2-9,15,20H,10-12,27H2,1H3,(H,28,30,31)/t15-,20-
InChIKey:
QKNBRNSGPNCARD-SGNKCFNYSA-N
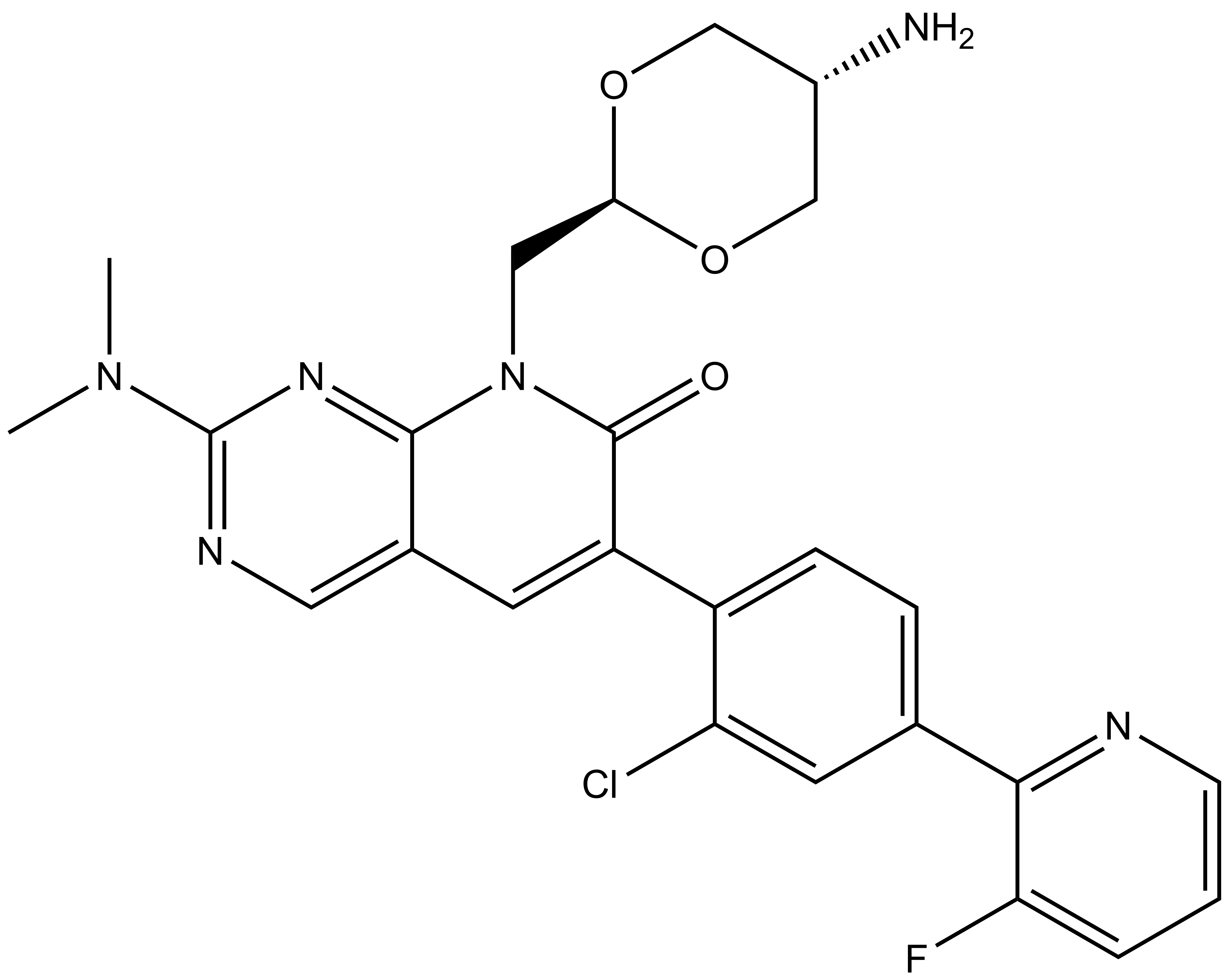
| Physical and chemical properties MR7 | |
| Molecular weight | 510.95 |
| Molecular formula | C25H24ClFN6O3 |
| IUPAC name | 8-(((2r,5r)-5-amino-1,3-dioxan-2-yl)methyl)-6-(2-chloro-4-(3-fluoropyridin-2-yl)phenyl)-2-(dimethylamino)pyrido[2,3-d]pyrimidin-7(8H)-one |
| logP | 2.95 |
| PSA | 105.11 |
| No. of chiral centre | 0 |
| No. of rotatable bonds | 8 |
| No. of hydrogen bond acceptors | 8 |
| No. of hydrogen bond donors | 1 |
| Storage | stable as solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | soluble in DMSO in a concentration of 50 mM |
SMILES:
CN(C1=NC=C2C(N(C(C(C3=CC=C(C=C3Cl)C4=NC=CC=C4F)=C2)=O)C[C@H]5OC[C@@H](CO5)N)=N1)C
InChI: InChI=1S/C25H24ClFN6O3/c1-32(2)25-30-10-15-8-18(17-6-5-14(9-19(17)26)22-20(27)4-3-7-29-22)24(34)33(23(15)31-25)11-21-35-12-16(28)13-36-21/h3-10,16,21H,11-13,28H2,1-2H3/t16-,21-
InChIKey:
RRFFBKGCCWPMLK-OQIWPSSASA-N
Selectivity profile of MRIA9 was determined with the 33PanQinase activity assay from Reaction Biology at 1uM and off targets were confirmed with in vitro IC50 with the same assay and in cellulo IC50 with NanoBRET™ assay. MRIA9 is a pan SIK and group I PAK inhibitor.
| Kinase | Percent of control(%) | 33 PanQinase IC 50 (nM) | NanoBRET IC 50 (nM) |
| SIK2 | 1 | 48 | 180 |
| SIK3 | 2 | 22 | 127 |
| SIK1 | 4 | 55 | 516 |
| KHS1 | 8 | 210 | 13000 |
| PAK3 | 9 | 140 | n.d |
| PAK2 | 10 | 41 | n.d |
| NLK | 13 | 250 | 3100 |
| PKN3 | 35 | 1400 | 6700 |
| PAK1 | 36 | 580 | n.d |
| MAP2K4 | 37 | 830 | n.d |
| TIE2 | 39 | 3100 | 6000 |
| MST4 | 45 | 1600 | 34000 |
| MELK | 48 | 2200 | n.d |
The negative control MR7 with its blocked hinge-binding amine showed no activity on a DSF assay for 100 kinases and low activity on target on NanoBRET assay.
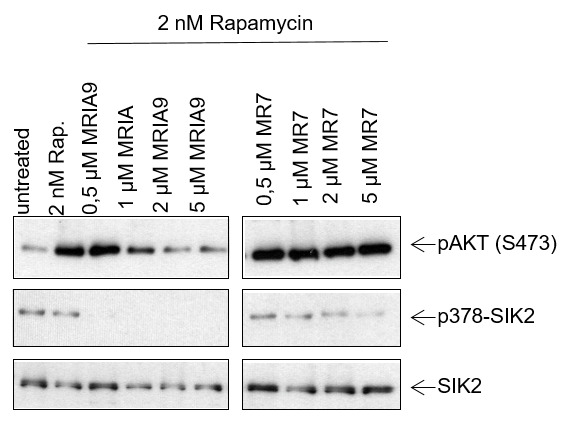
 |
MRIA9 modulated endogenous substrates linked to SIK activity. In the ovarian cancer cell SKOV-3 in which the PI3K/AKT/MTOR pathway was activated by rapamycine, MRIA9 abrogated the phosphorylation of AKT in a dose-dependent manner. In addition, SIK2 auto-phosphorylation activity was completely inhibited whereas the negative control did not influence SIK2 activity.
| SKOV-3 cell line |
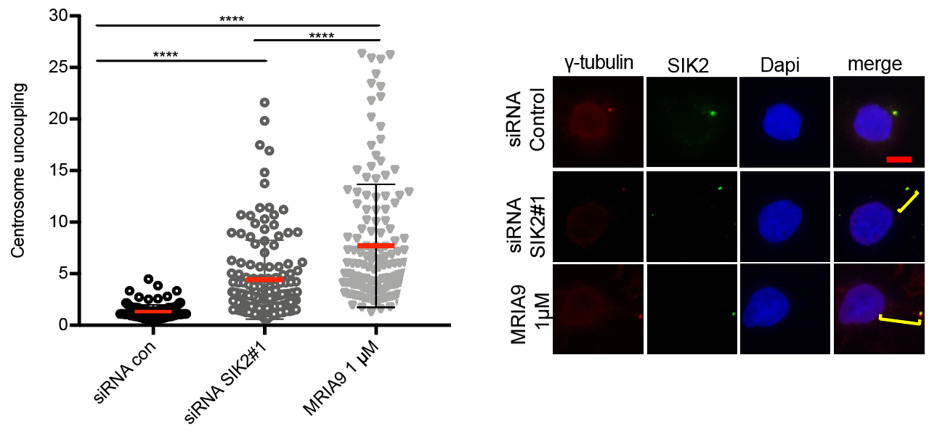
 |
MRIA9 replicated a known phenotype of SIK inhibition, displacing the centrosome from the nucleous in ovarian cancer cell line SKOV-3, similar to the phenotype seen when silence RNA is used to knock down SIK2.
| SKOV-3 cell line |
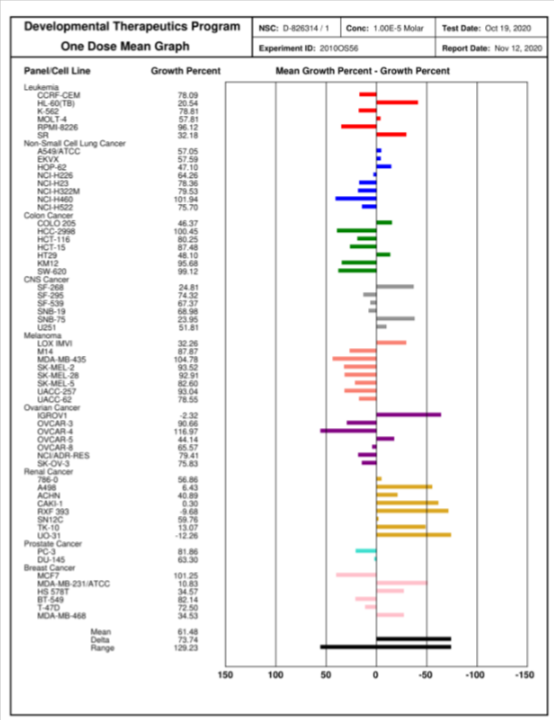
 |
In the NCI-60 screen, which is a human tumor cell line screen, MRIA9 showed only modest cell toxicity or growth inhibition. It was tested in a single high dose of 10 µM in the full NCI-60 panel. https://dtp.cancer.gov/discovery_development/nci-60/methodology.htm
(1) Sun, Z.; Jiang, Q.; Li, J.; Guo, J. The potent roles of salt-inducible kinases (SIKs) in metabolic homeostasis and tumorigenesis. Signal transduction and targeted therapy 2020, 5 (1), 150.
(2) Conkright, M. D.; Canettieri, G.; Screaton, R.; Guzman, E.; Miraglia, L.; Hogenesch, J. B.; Montminy, M. TORCs: transducers of regulated CREB activity. Molecular cell 2003, 12 (2), 413–423.
(3) Katoh, Y.; Takemori, H.; Lin, X.-Z.; Tamura, M.; Muraoka, M.; Satoh, T.; Tsuchiya, Y.; Min, L.; Doi, J.; Miyauchi, A.; Witters, L. A.; Nakamura, H.; Okamoto, M. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. The FEBS journal 2006, 273 (12), 2730–2748.
(4) Chen, F.; Chen, L.; Qin, Q.; Sun, X. Salt-inducible kinase 2: an oncogenic signal transmitter and potential target for cancer therapy. Frontiers in oncology 2019, 9, 18.
(5) Cheng, H.; Liu, P.; Wang, Z. C.; Zou, L.; Santiago, S.; Garbitt, V.; Gjoerup, O. V.; Iglehart, J. D.; Miron, A.; Richardson, A. L.; Hahn, W. C.; Zhao, J. J. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Science signaling 2009, 2 (80), ra35.
(6) Montenegro, R. C.; Howarth, A.; Ceroni, A.; Fedele, V.; Farran, B.; Mesquita, F. P.; Frejno, M.; Berger, B.-T.; Heinzlmeir, S.; Sailem, H. Z.; Tesch, R.; Ebner, D.; Knapp, S.; Burbano, R.; Kuster, B.; Müller, S. Identification of molecular targets for the targeted treatment of gastric cancer using dasatinib. Oncotarget 2020, 11 (5), 535–549.
(7) Tesch, R.; Rak, M.; Raab, M.; Berger, L. M.; Kronenberger, T.; Joerger, A. C.; Berger, B.-T.; Abdi, I.; Hanke, T.; Poso, A.; Strebhardt, K.; Sanhaji, M.; Knapp, S. Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors. Journal of Medicinal Chemistry 2021, 64 (12), 8142-8160.
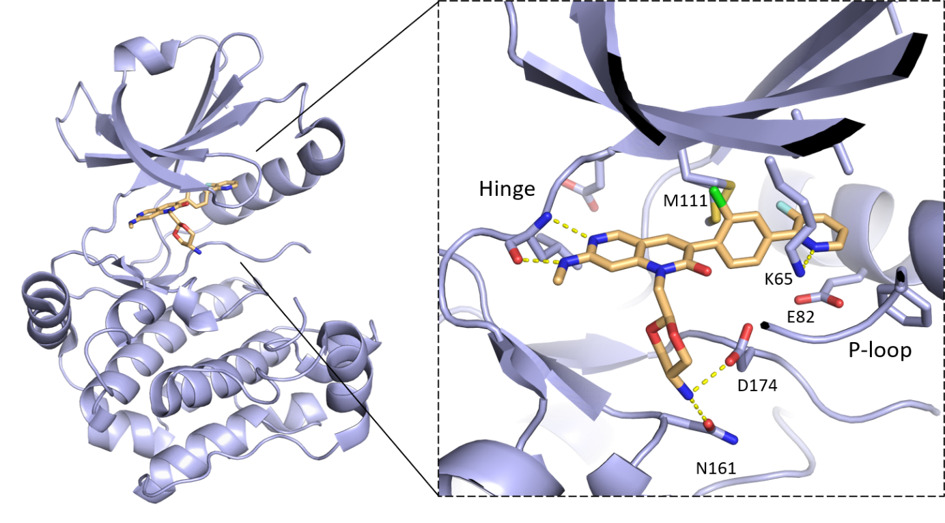
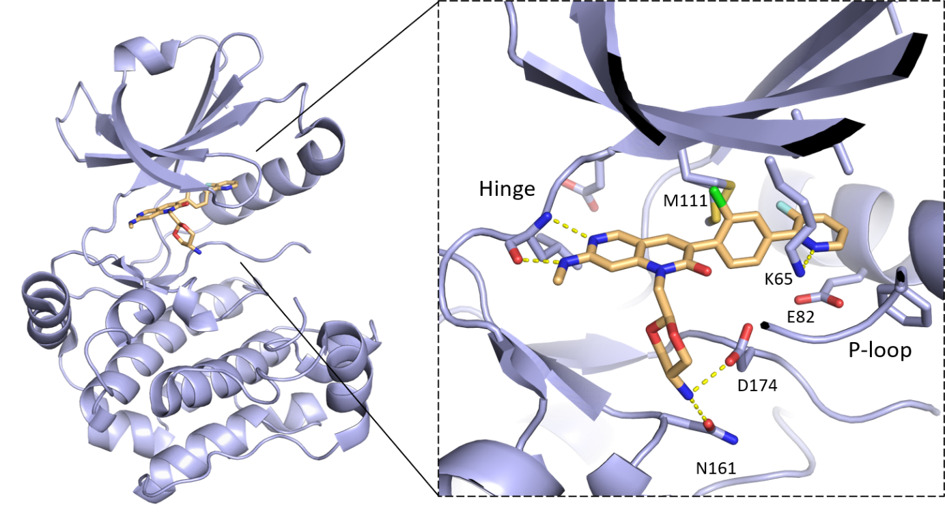
Binding mode of MRIA9 in complex with the crystallographic surrogate model MST3 (PDB 7B31). The inhibitor binds to the ATP pocket and different stability on the P-loop region of MST3 and SIK2 studied by molecular dynamics, explain the selectivity of MRIA9 towards SIK2. [7]