The probe NVS-BPTF-1 is available from Cayman Chemical.
| Probe | Negative control | |
 |
|  |
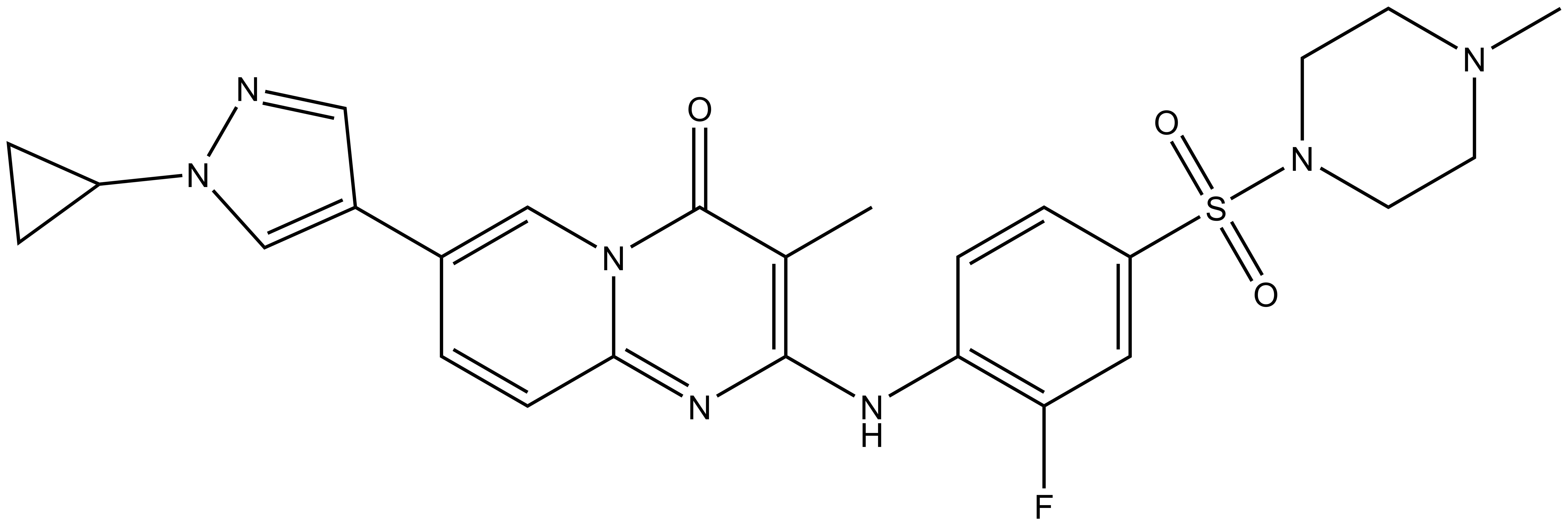
NVS-BPTF-1 |
| NVS-BPTF-C |
BPTF (Bromodomain PHD-finer Transcription Factor, also known as FALZ) is a multi-domain protein containing multiple histone lysine recognition domains. BPTF binds to acetylated H3 and H4 histone tails via its Bromodomain and H3K4me3 via the second PHD domain (1). BPTF is part of the NURF chromatin remodelling complex and involved in maintaining chromatin accessibility (2).
BPTF has been shown to be essential for normal embryogenesis (3) with studies indicating key roles in controlling embryonic stem cell differentiation (4). Recently activation of BPTF expression by MITF (Microphthalmia-associated Transcription Factor) has been linked to transducing key pro-survival signals in melanoma (5). Small molecule inhibition of BPTF may therefore be a viable approach for the treatment of melanoma and other malignancies.
Novartis in partnership with the SGC has developed NVS-BPTF-1 as a potent, selective and cell active chemical probe for BPTF. NVS-BPTF-1 and its structurally very similar control NVS-BPTF-C will assist scientists in further dissecting the key role that BPTF1 plays in multiple signaling pathways for cell proliferation and survival.
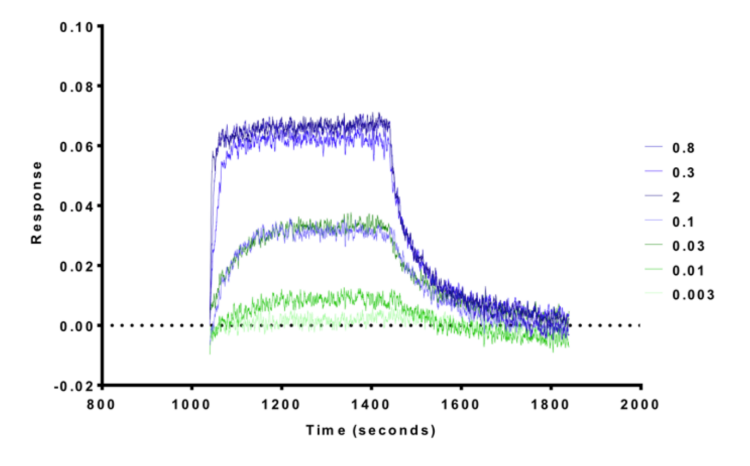
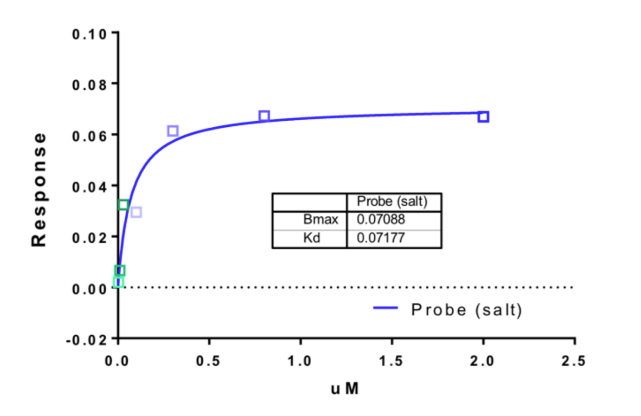
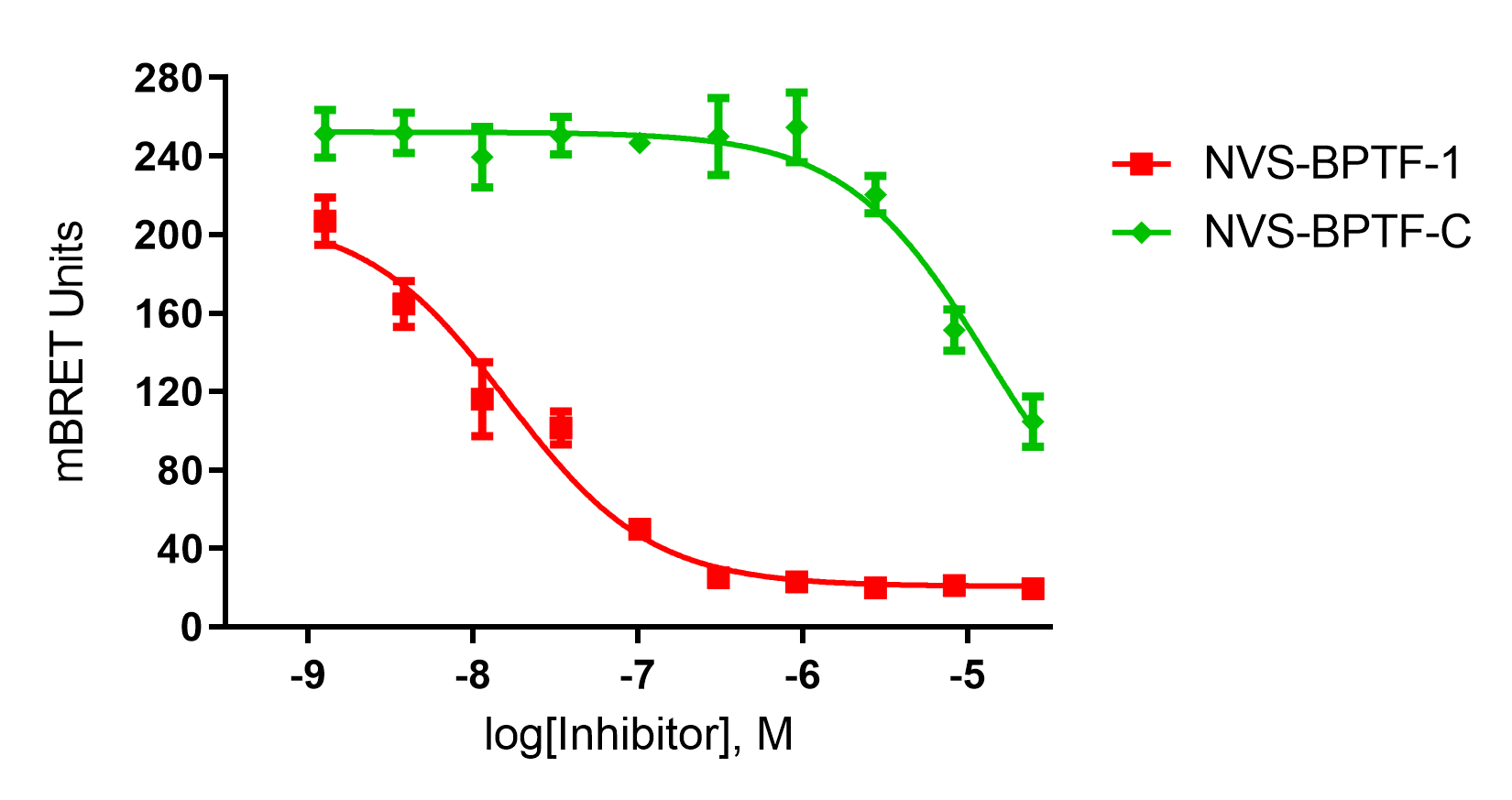
NVS-BPTF-1 has on target in vitro binding affinity of 71nM with BPTF. The negative control NVS-BPTF-C shows a binding affinity of 1.67µM with BPTF.
In HEK293 cells, NVS-BPTF-1 exhibits an on-target IC50 of 16nM (nanoBRET) whilst the negative control NVS-BPTF-C shows no activity.
Note that NVS-BPTF-1 does not have adequate ADME properties for in vivo work.
NVS-BPTF-1 is poorly soluble except in the HCl salt form. High cellular potency enables low concentrations of the compound to be used in cellular applications. It is recommended that the compounds are used at concentrations below 1µM.
NVS-BPTF-1 shows an IC50 of 56nM against BPTF in an alphascreen assay and a KD of 71nM in a BLI assay.
| Probe |
 |
NVS-BPTF-1 |
| Physical and chemical properties for NVS-BPTF-1 | |
| Molecular weight | 537.19 |
| Molecular formula | C26H28FN7O3S |
| IUPAC Name | 9-(1-cyclopropyl-1H-pyrazol-4-yl)-4-(2-fluoro-4-(4-methyl-piperazin-1-ylsulfonyl)-phenylamino)-3-methyl-1,5-diaza-bicyclo[4.4.0]deca-3,5,7,9-tetraen-2-one |
| MollogP | 2.9 |
| PSA | 82.9 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 10 |
| No. of hydrogen bond donors | 1 |
| Permeability of HCl Salt | PAMPA: Log PAMPA (cm/s): -4.8 Calculate %FA: 69 Class: medium MDCK low efflux: 20% of recovery |
| Solubility | Equilibrium solubility: pH 4(g/L): >0.538 pH 6.8 (g/L): <0.002 Thermodynamic solubility: pH 1 (g/L): 0.074 pH 6.8 (g/L): 0.0008 |
| Storage | Store at -20°C |
| Dissolution | Up to 50 mM in DMSO |
| Negative control |
 |
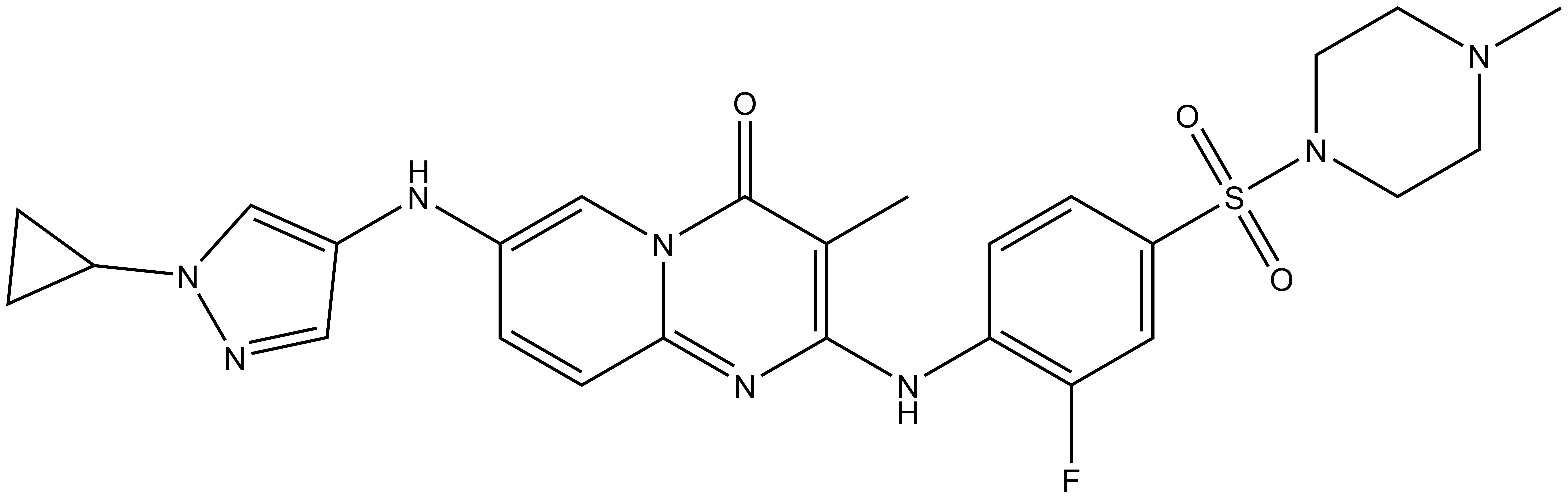
NVS-BPTF-C |
| Physical and chemical properties for NVS-BPTF-C | |
| Molecular weight | 552.21 |
| Molecular formula | C26H29FN8O3S |
| IUPAC Name | 9-(1-cyclopropyl-1H-pyrazol-4-ylamino)-4-(2-fluoro-4-(4-methyl-piperazin-1-ylsulfonyl)-phenylamino)-3-methyl-1,5-diaza-bicyclo[4.4.0]deca-3,5,7,9-tetraen-2-one |
| MollogP | 2.43 |
| PSA | 92.7 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 7 |
| No. of hydrogen bond acceptors | 10 |
| No. of hydrogen bond donors | 2 |
| Storage | Store at -20°C |
| Dissolution | Up to 50 mM in DMSO |
SMILES:
NVS-BPTF-1: CC1=C(N=C2C=CC(C3=CN(C4CC4)N=C3)=CN2C1=O)NC5=C(F)C=C(S(N6CCN(CC6)C)(=O)=O)C=C5
NVS-BPTF-C: CC1=C(N=C2C=CC(NC3=CN(C4CC4)N=C3)=CN2C1=O)NC5=C(F)C=C(S(N6CCN(CC6)C)(=O)=O)C=C5
InChI:
NVS-BPTF-1: InChI=1S/C26H28FN7O3S/c1-17-25(29-23-7-6-21(13-22(23)27)38(36,37)32-11-9-31(2)10-12-32)30-24-8-3-18(15-33(24)26(17)35)19-14-28-34(16-19)20-4-5-20/h3,6-8,13-16,20,29H,4-5,9-12H2,1-2H3
NVS-BPTF-C: InChI=1S/C26H29FN8O3S/c1-17-25(30-23-7-6-21(13-22(23)27)39(37,38)33-11-9-32(2)10-12-33)31-24-8-3-18(15-34(24)26(17)36)29-19-14-28-35(16-19)20-4-5-20/h3,6-8,13-16,20,29-30H,4-5,9-12H2,1-2H3
InChIKey:
NVS-BPTF-1: JYTISQGEFSHUIR-UHFFFAOYSA-N
NVS-BPTF-C: BFSKPRUNBBMNCF-UHFFFAOYSA-N
A DSF screen against Human bromodomains reveals no significant off-targets (Table 1). A BROMOscan with NVS-BPTF-1 similarly showed good selectivity. BPTF exhibited a KD of 3nM, BRPF 37nM, CECR2 66nM, GCN5L2 62nM and PCAF 74nM in BROMOscan (Table 2)
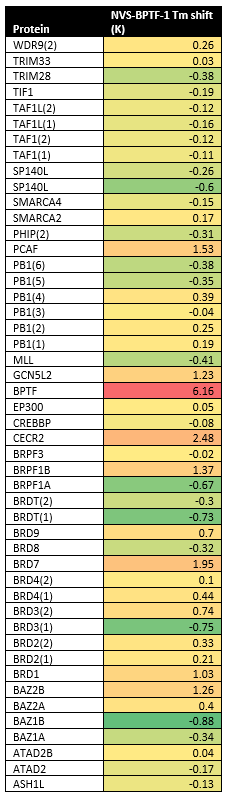
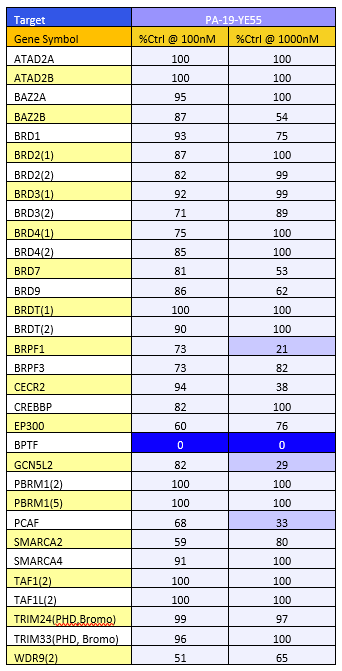
Table 1: DSF screen Table 2: BromoScan
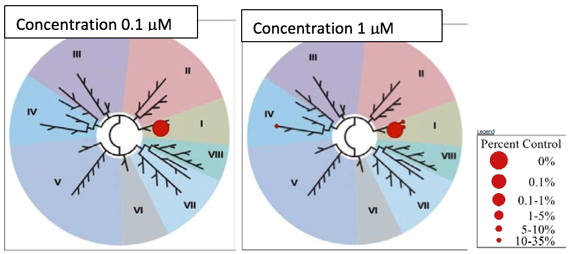
NVS-BPTF-1 was also tested against the NIBR principal panel which showed no binding to 12 GPCRs, 3 nuclear receptors, 3 transporters and 7 other enzymes with an IC50<10µM.

The control compound, NVS-BPTF-C showed no binding to 14 GPCRs, 3 nuclear receptors, 3 transporters and 5 enzymes with an IC50<10µM in the NIBR principal panel.

Against the NIBR kinase panel, NVS-BPTF-1 showed no binding against 48 kinases with an IC50<30µM.
Against the NIBR kinase panel, NVS-BPTF-C showed no binding against 59 kinases with an IC50<30µM.
Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Agilent). Proteins were prepared in 10 mM HEPES pH 7.5, 500 mM NaCl and assayed in a 96-well plate at a final concentration of 2 μM in 20 μL volume. Compounds final concentration is 10 μM.
All bromodomain proteins were prepared according to the published procedures (Filippakopoulos at al, 2012). Assay was performed as described previously (Philpott et al, 2011). All reagents were pre-diluted in 25 mM HEPES, 100 mM NaCl, 0.1 % BSA, pH 7.4 and 0.05 % CHAPS and allowed to equilibrate to room temperature prior to addition to plates. Plates filled with 5 uL of the assay buffer followed by 7 µL of biotinylated peptide [H-YSGRGKacGGKacGLGKacGGAKacRHRK(Biotin)-OH and His-tagged protein to achieve final assay concentrations of 25 nM. Plates were sealed and incubated for a further 60 minutes, before the addition of 8 μl of the mixture of streptavidin-coated donor beads (12.5 μg/ml) and nickel chelate acceptor beads (12.5 μg/ml) under low light conditions. Plates were foil-sealed to protect from light, incubated at room temperature for 60 minutes and read on a PHERAstar FS plate reader (BMG Labtech, Germany) using an AlphaScreen 680 excitation/570 emission filter set.
BioLayer Interferometry (BLI) experiments were performed on a 16-channel ForteBio Octet RED384 instrument at 25 °C in 25mM HEPES, pH 7.5, and 100mM NaCl buffer. Proteins were biotinylated in vivo using the construct with C-terminal AviTag. Proteins were immobilized on SSA sensors. Measurements were performed using 240 sec association step followed by 240 sec dissociation step on black tilted low volume 384-well plate (ForteBio). Baseline was stabilized for 120 sec prior to association step. Signal from the reference sensors was subtracted prior to Kd calculations using Analysis software (ForteBio).
To determine cellular target engagement, a NanoBRET assay based on a bespoke tracer (5961) was developed to measure the binding of the chemical probe and negative control to the NanoLuc-tagged BPTF bromodomain.
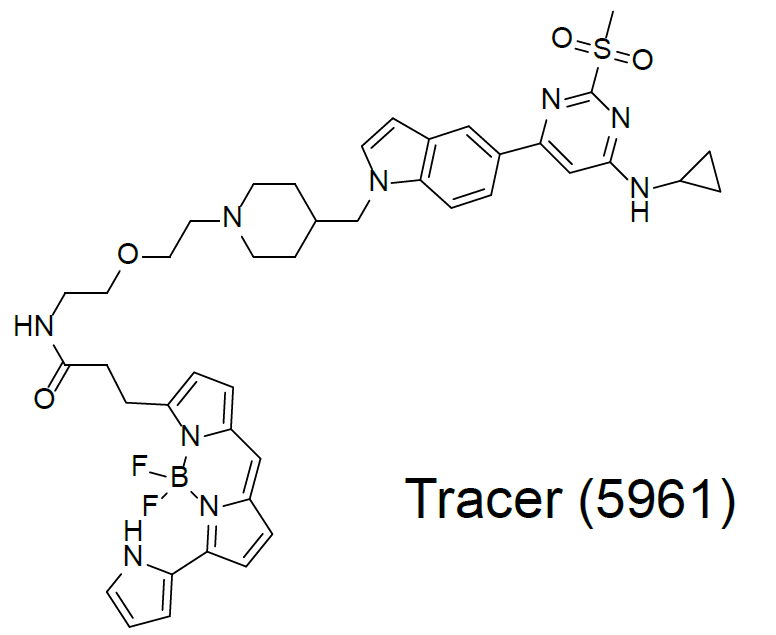
An EC50 of 16 nM was observed in this assay for NVS-BPTF-1 whereas the negative control NVS-BPTF-C was significantly less potent.
The putative cellular off-targets CECR2, PCAF and GCN5L2 were similarly tested in the NanoBRET assay with no activity detected.
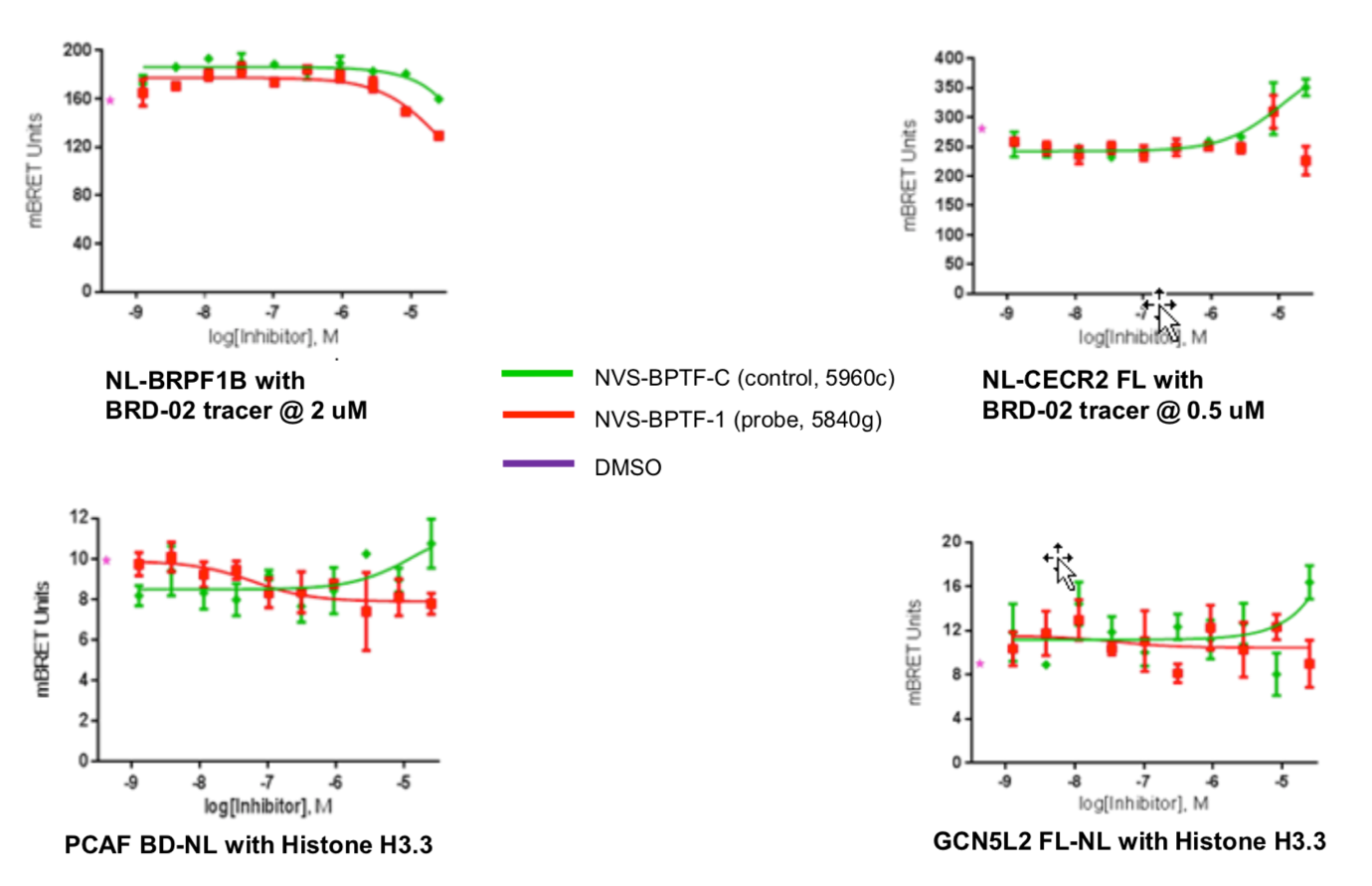
Dose-response experiments were conducted in 384 well format using HEK293T cells expressing NanoLuc fused to the N-terminus of the BPTF bromodomain, using the 5961 tracer at a final concentration of 1 µM.
1. Wysocka, J., Swigut, T., Xiao, H., Milne, T. A., Kwon, S. Y., Landry, J., Kauer, M., Tackett, A. J., Chait, B. T., Badenhorst, P., Wu, C., and Allis, C. D. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86-90
2. Goller, T., Vauti, F., Ramasamy, S., and Arnold, H. H. (2008) Transcriptional regulator BPTF/FAC1 is essential for trophoblast differentiation during early mouse development. Molecular and cellular biology 28, 6819-6827
3. Landry, J., Sharov, A. A., Piao, Y., Sharova, L. V., Xiao, H., Southon, E., Matta, J., Tessarollo, L., Zhang, Y. E., Ko, M. S., Kuehn, M. R., Yamaguchi, T. P., and Wu, C. (2008) Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS genetics 4, e1000241
4. Ngeow, K. C., Friedrichsen, H. J., Li, L., Zeng, Z., Andrews, S., Volpon, L., Brunsdon, H., Berridge, G., Picaud, S., Fischer, R., Lisle, R., Knapp, S., Filippakopoulos, P., Knowles, H., Steingrimsson, E., Borden, K. L. B., Patton, E. E., and Goding, C. R. (2018) BRAF/MAPK and GSK3 signaling converges to control MITF nuclear export. Proceedings of the National Academy of Sciences of the United States of America 115, E8668-e8677
5. Dar, A. A.; Majid, S.; Bezrookove, V.; Phan, B.; Ursu, S.; Nosrati, M.; De Semir, D.; Sagebiel, R. W.; Miller, J. R.; Debs, R.; Cleaver, J. E.; Kashani-Sabet, M., BPTF transduces MITF-driven prosurvival signals in melanoma cells. Proc. Natl. Acad. Sci. U. S. A., 2016, 113 (22), 6254–6258.
Novartis have solved the structure of NVS-BPTF-1 in complex with BPTF at a resolution of 1.76Å. This structure is not currently deposited in the PDB.
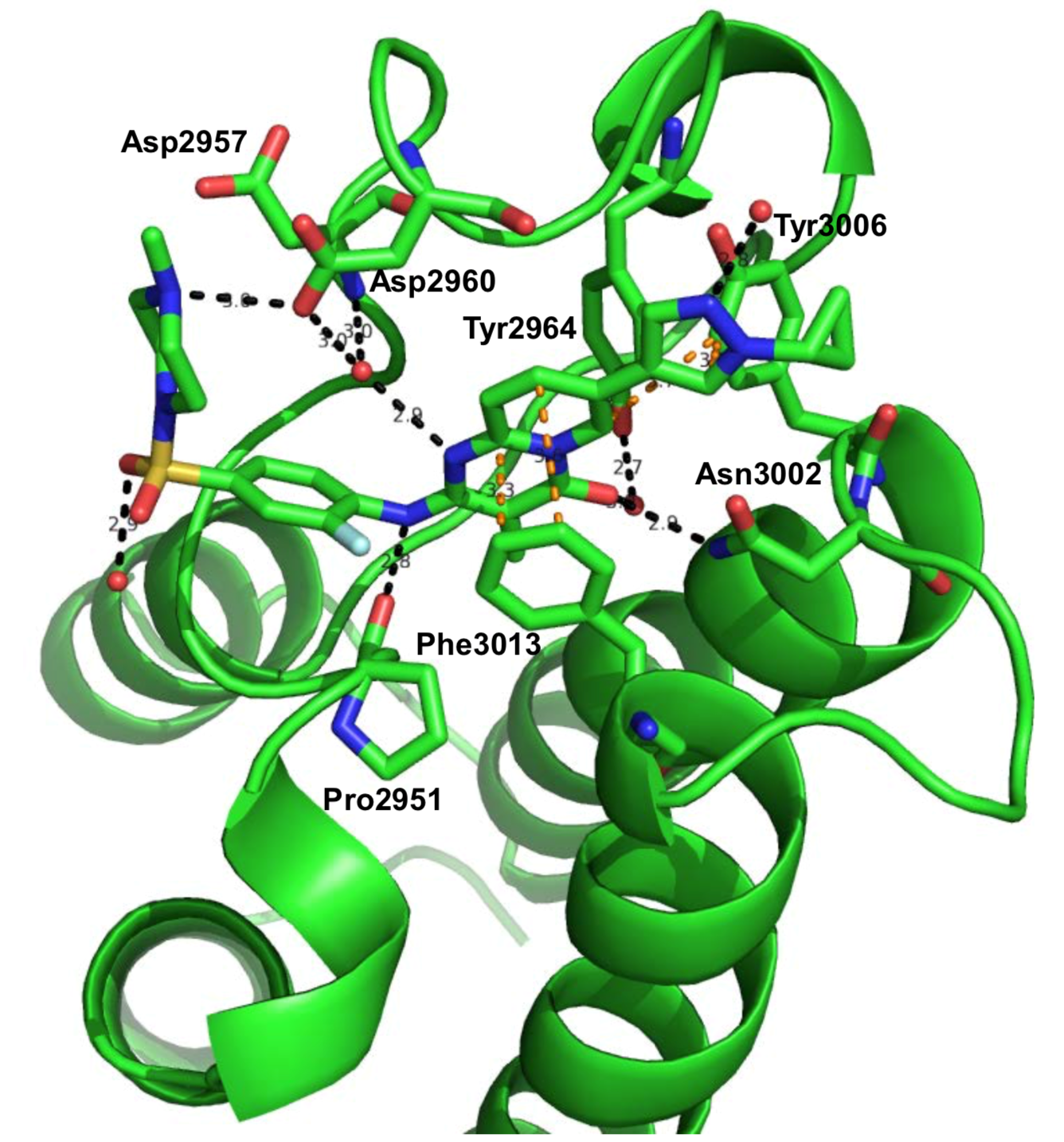
Black dotted-line: H-Bond
Orange dotted-line: pi-pi interaction
Red sphere: water molecule