This probe is available from Sigma.
| Probe | Negative control | |
 |
|  |
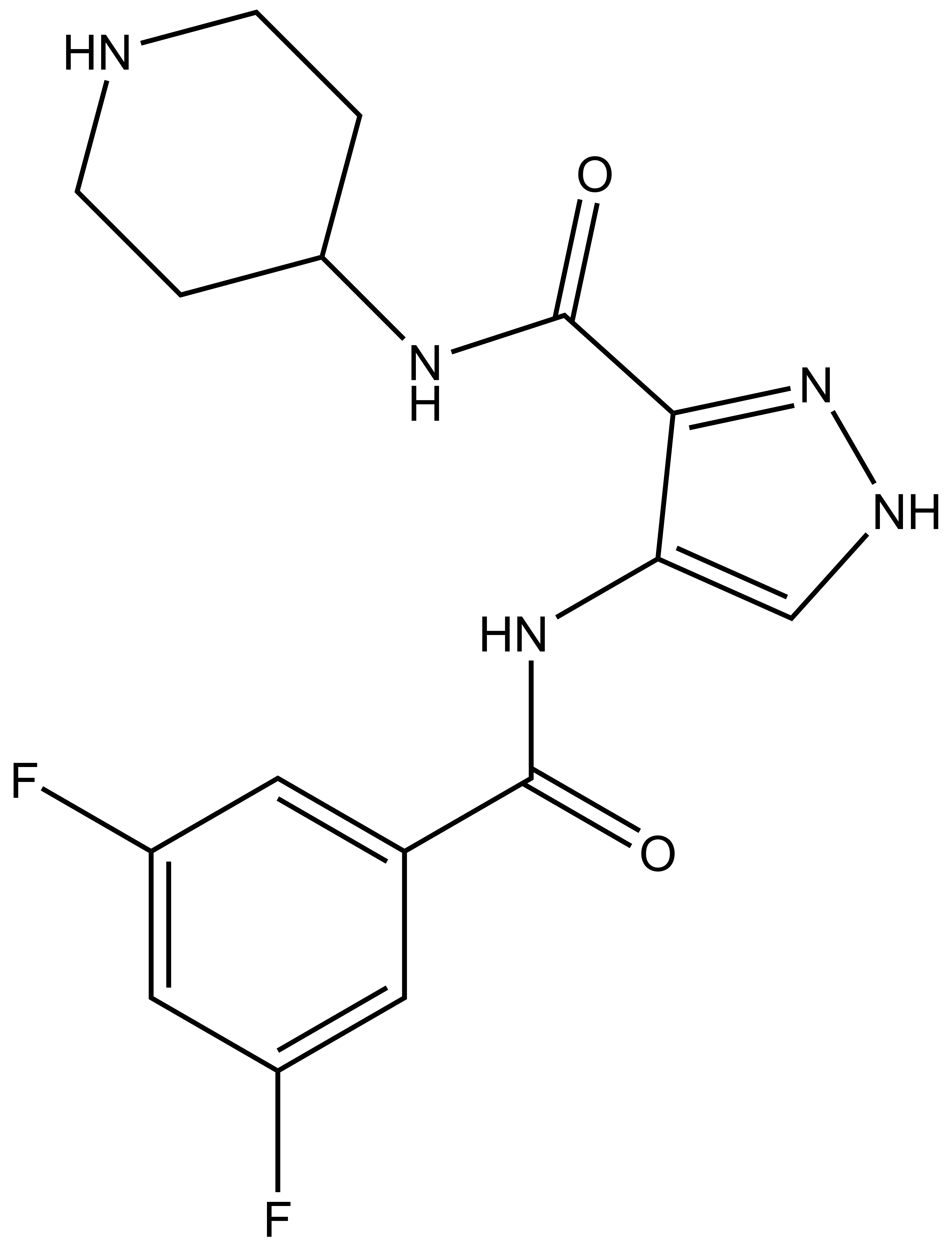
SGC-CDKL5/GSK3-1 |
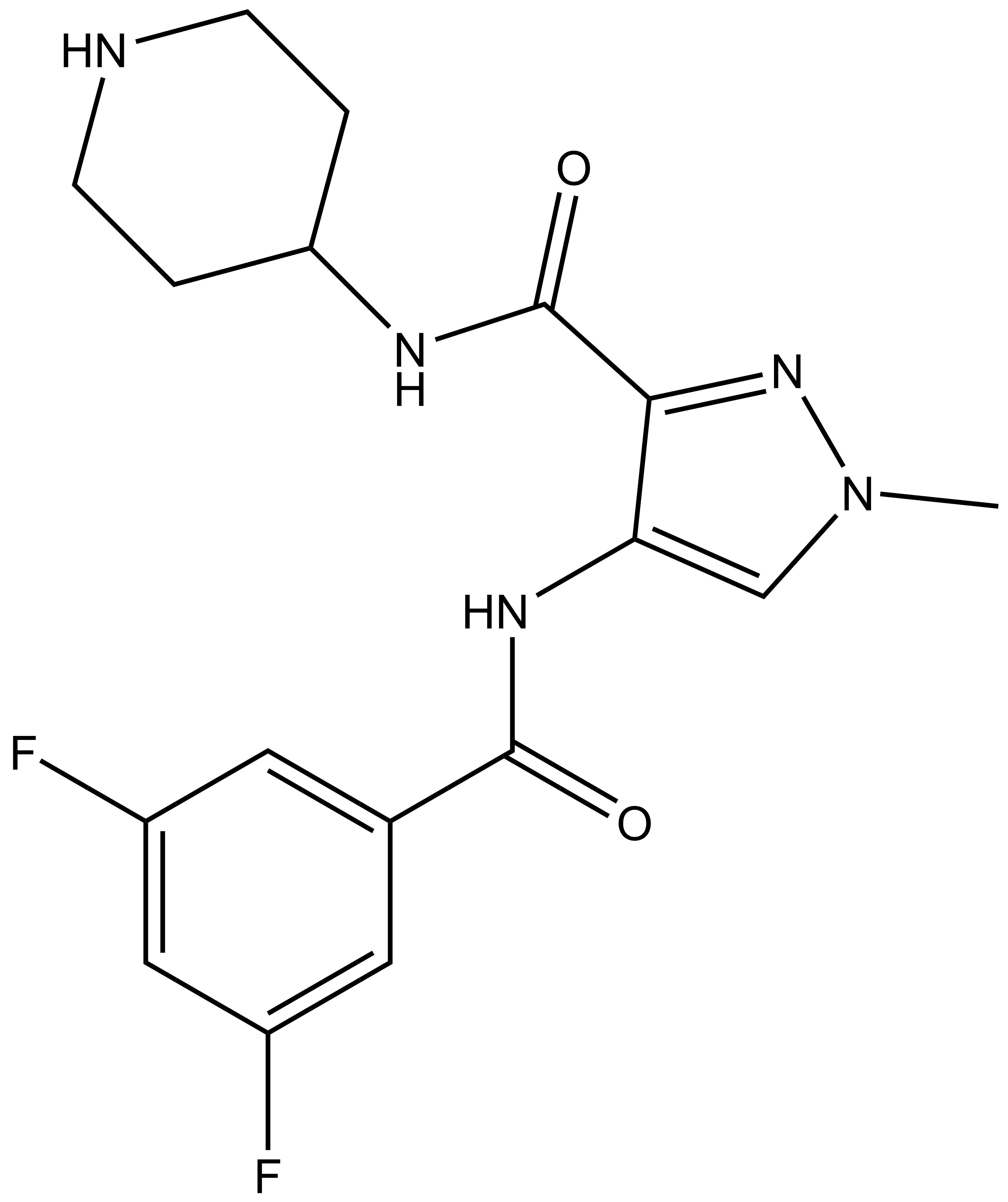
| SGC-CDKL5/GSK3-1N |
From a library of AT-7519 analogs, we identified a potent and cell-active chemical probe (SGC-CDKL5/GSK3-1) that inhibits cyclin-dependent kinase-like 5 (CDKL5) and glycogen synthase kinase-3 (GSK3⍺ and β). Comprehensive evaluation of kinome-wide selectivity confirmed that this CDKL5/GSK3 probe demonstrates exceptional selectivity. A structurally similar analog (SGC-CDKL5/GSK3-1N) was characterized as a negative control that does not bind to CDKL5, GSK3⍺, or GSK3β in corresponding cellular target engagement assays. At nanomolar concentrations, our CDKL5/GSK3 chemical probe promoted motor neuron survival when iPSC-derived motor neurons were subjected to ER stress. We recommend use of a structurally related GSK3 chemical probe, SGC-GSK3-1, in tandem to interrogate those effects due to GSK3 rather than CDKL5 inhibition. An orthogonal CDKL5 chemical probe, SGC-CAF382-1, represents a distinct chemotype with non-overlapping kinase off-targets that can also be used in parallel. Our chemical probe set can be used by the community to further characterize the underexplored roles of CDKL5 and how inhibition of CDKL5 and GSK3 impacts downstream biology.
| Probe | Negative control | |
 |
|  |
SGC-CDKL5/GSK3-1 |
| SGC-CDKL5/GSK3-1N |
| Physical and chemical properties for SGC-CDKL5/GSK3-1 | |
| Molecular weight | 349.34 |
| Molecular formula | C16H17FN5O2 |
| IUPAC name | 4-(3,5-difluorobenzamido)-N-(piperidin-4-yl)-1H-pyrazole-3-carboxamide |
| ClogP | -0.33 |
| PSA | 98.91 |
| No. of chiral centers | 0 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 4 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10 mM) are stable at -20oC |
| Dissolution | Soluble in DMSO up to 10 mM |
| Physical and chemical properties for SGC-CDKL5/GSK3-1N | |
| Molecular weight | 363.37 |
| Molecular formula | C17H19FN5O2 |
| IUPAC name | 4-(3,5-difluorobenzamido)-1-methyl-N-(piperidin-4-yl)-1H-pyrazole-3-carboxamide |
| ClogP | -0.32 |
| PSA | 88.05 |
| No. of chiral centers | 0 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 3 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10 mM) are stable at -20oC |
| Dissolution | Soluble in DMSO up to 10 mM |
SMILES:
SGC-CDKL5/GSK3-1: O=C(NC1CCNCC1)C2=NNC=C2NC(C3=CC(F)=CC(F)=C3)=O
SGC-CDKL5/GSK3-1N: O=C(NC1CCNCC1)C2=NN(C=C2NC(C3=CC(F)=CC(F)=C3)=O)C
InChI:
SGC-CDKL5/GSK3-1: InChI=1S/C16H17F2N5O2/c17-10-5-9(6-11(18)7-10)15(24)22-13-8-20-23-14(13)16(25)21-12-1-3-19-4-2-12/h5-8,12,19H,1-4H2,(H,20,23)(H,21,25)(H,22,24)
SGC-CDKL5/GSK3-1N: InChI=1S/C17H19F2N5O2/c1-24-9-14(22-16(25)10-6-11(18)8-12(19)7-10)15(23-24)17(26)21-13-2-4-20-5-3-13/h6-9,13,20H,2-5H2,1H3,(H,21,26)(H,22,25)
InChIKey:
SGC-CDKL5/GSK3-1: NIHAFOURWLZLFN-UHFFFAOYSA-N
SGC-CDKL5/GSK3-1N: ODZBDODDFLVDQZ-UHFFFAOYSA-N
SGC-CDKL5/GSK3-1 was profiled in the DiscoverX scanMAX assay against 403 wild-type kinases at 1 μM. Only 4 kinases showed PoC <10 giving an S10(1 μM) = 0.01. When the PoC <35 fraction was examined, 11 kinases were included (S35(1 μM) = 0.027). Potential off-targets within the S35(1 μM) fraction were tested via biochemical enzymatic/binding assays plus NanoBRET target engagement assays for CDKL5, GSK3⍺, GSK3β, CDK16, and CDK17. Data corresponding with off-target kinase activity is shown in the table below.
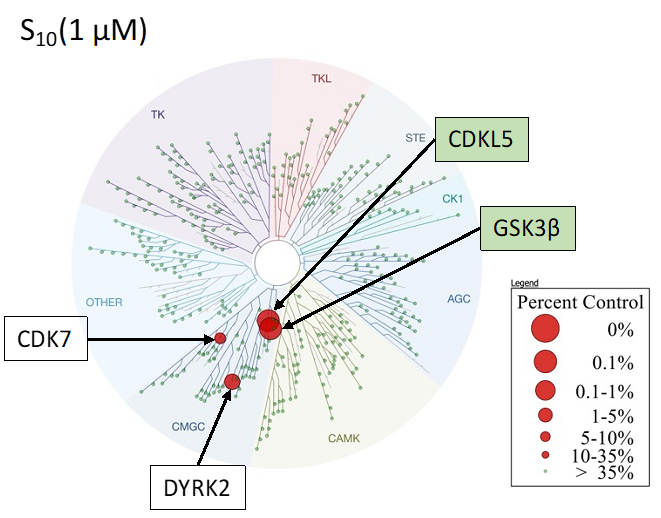
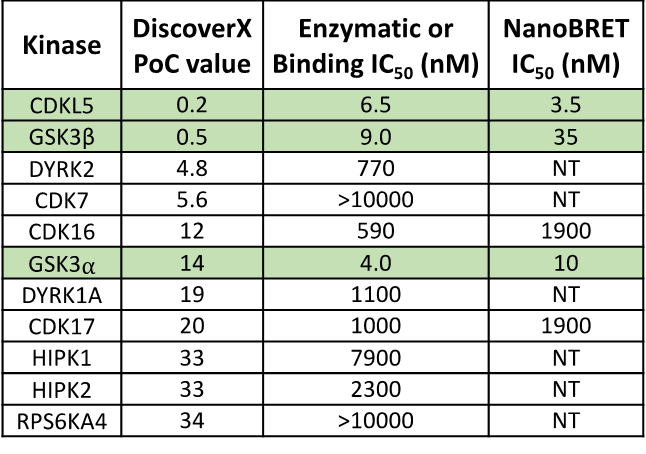
Figure 2: SGC-CDKL5/GSK3-1 was profiled in the DiscoverX scanMAX assay against 403 wild-type kinases at 1 μM and off-target kinases inhibited PoC <35 were tested in an orthogonal assay. Rows colored green are CDKL5, GSK3⍺, and GSK3β. No other kinases demonstrate enzymatic IC50 values within 30-fold of the GSK3β enzymatic IC50 value.
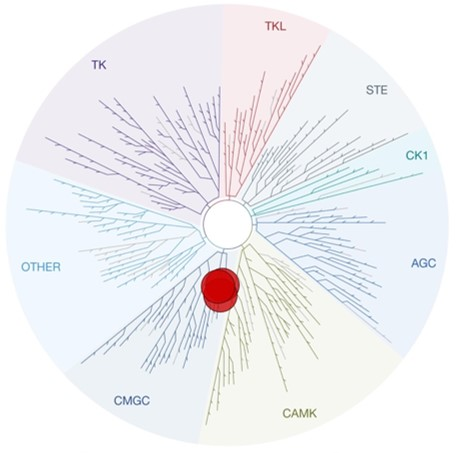
Figure 1: Kinome tree with CDKL5, GSK3⍺ and GSK3β highlighted as red circles. Illustration is reproduced courtesy of Eurofins DiscoverX (http://treespot.discoverx.com).
Biological activity summary:
A NanoBRET assay was utilized to assess the binding affinity of SGC-CDKL5/GSK3-1 to CDKL5, GSK3⍺, GSK3β, CDK16, and CDK17. The negative control shows no binding affinity for CDKL5, GSK3⍺, or GSK3β.
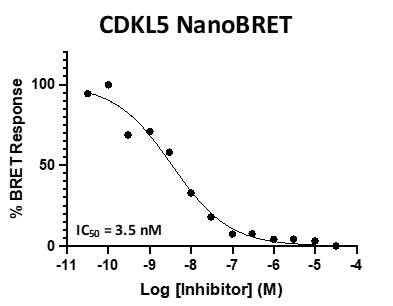
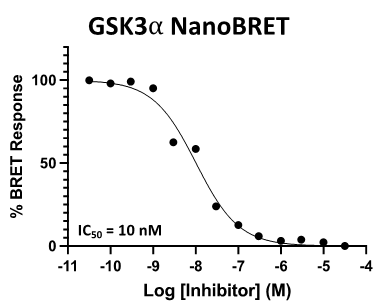
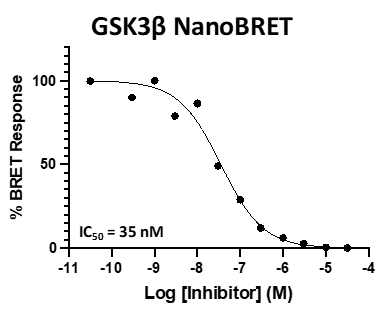
Figure 3: SGC-CDKL5/GSK3-1 was profiled in the CDKL5, GSK3⍺, and GSK3β NanoBRET assays.
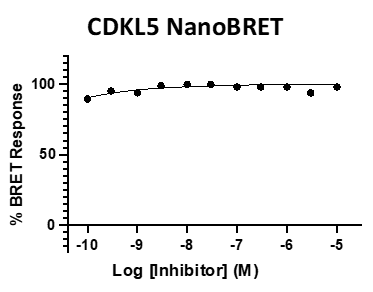
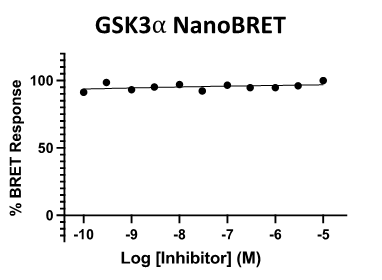

Figure 4: SGC-CDKL5/GSK3-1N was profiled in the CDKL5, GSK3⍺, and GSK3β NanoBRET assays.
References
Ong, H. W.; Liang, Y.; Richardson, W.; Lowry, E. R.; Wells, C. I.; Chen, X.; Silvestre, M.; Dempster, K.; Silvaroli, J. A.; Smith, J. L.; Wichterle, H.; Pabla, N. S.; Ultanir, S. K.; Bullock, A. N.; Drewry, D. H.*; Axtman, A. D.* Discovery of a potent and selective CDKL5/GSK3 chemical probe that is neuroprotective. ACS Chem Neurosci 2023, 14, 1672–1685; 10.1021/acschemneuro.3c00135.
Ong, H. W.; Liang, Y.; Richardson, W.; Lowry, E. R.; Wells, C. I.; Chen, X.; Silvestre, M.; Dempster, K.; Silvaroli, J. A.; Smith, J. L.; Wichterle, H.; Pabla, N. S.; Ultanir, S. K.; Bullock, A. N.; Drewry, D. H.*; Axtman, A. D.* A potent and selective CDKL5/GSK3 chemical probe is neuroprotective. BioRxiv 2023, doi: 10.1101/2023.02.09.527935.