The probe and control can be requested by clicking here.
| Probe | Negative control | |
 |
|  |
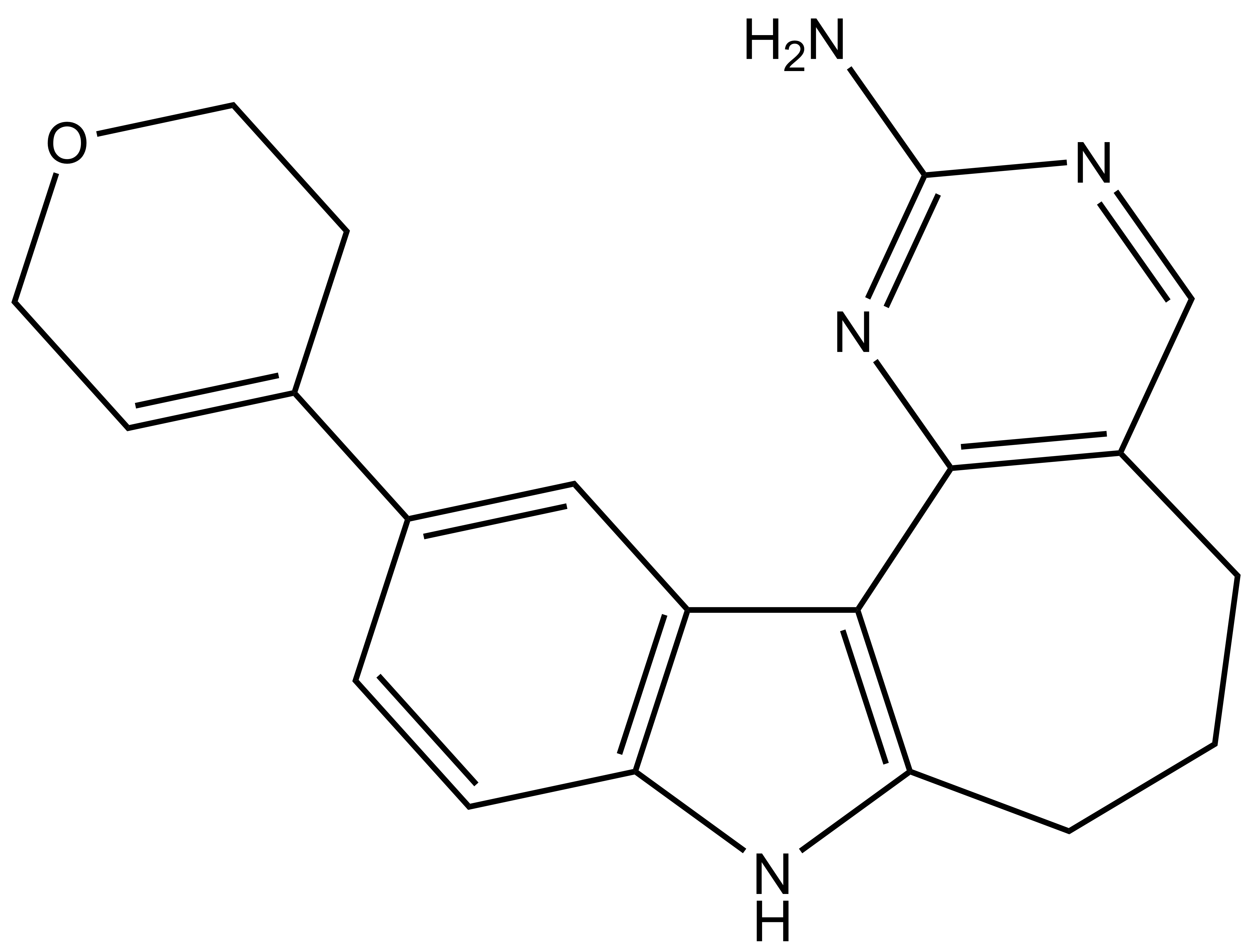
SGC-PI5P4Kγ/MYLK4-1 |
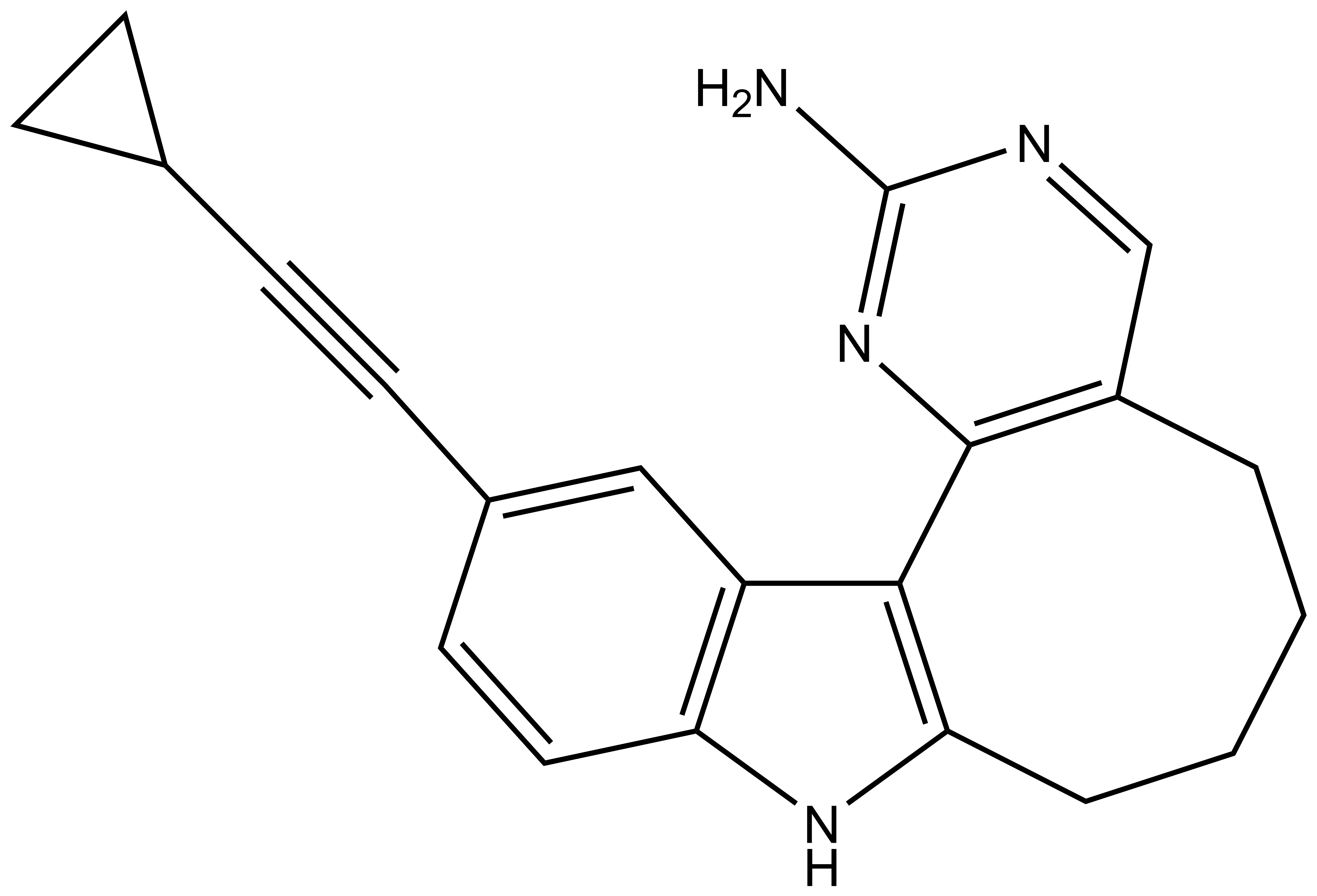
| SGC-PI5P4Kγ/MYLK4-1N |
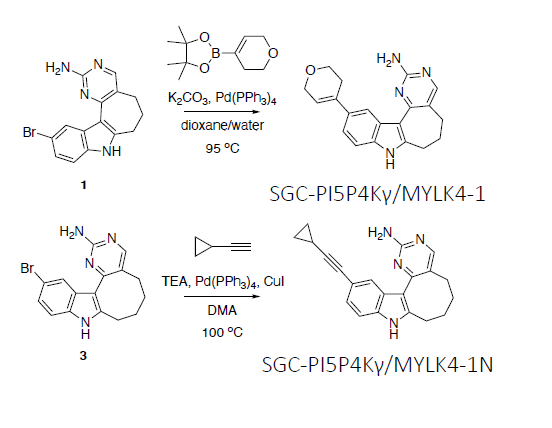
From a library of indolyl pyrimidinamines, we identified a potent and cell-active chemical probe (SGC-PI5P4Kγ/MYLK4-1) that inhibits phosphatidylinositol-5-phosphate 4-kinase gamma (PI5P4Kγ) and myosin light chain kinase family member 4 (MYLK4). Comprehensive evaluation of kinome-wide selectivity confirmed that this chemical probe demonstrates excellent selectivity. A structurally similar indolyl pyrimidinamine (SGC-PI5P4Kγ/MYLK4-1N) was characterized as a negative control that does not inhibit PI5P4Kγ or MYLK4 and exhibits exceptional selectivity when profiled broadly. Our PI5P4Kγ/MYLK4 chemical probe increases mTORC1 signaling in MCF-7 cells without associated toxicity. Our chemical probe set can be used by the community to further explore the biology regulated by PI5P4Kγ and/or MYLK4.
| Probe | Negative control | |
 |
|  |
SGC-PI5P4Kγ/MYLK4-1 |
| SGC-PI5P4Kγ/MYLK4-1N |
SMILES:
SGC-PI5P4Kγ/MYLK4-1: NC1=NC2=C(C=N1)CCCC3=C2C4=C(N3)C=CC(C5=CCOCC5)=C4
SGC-PI5P4Kγ/MYLK4-1N: NC1=NC2=C(C=N1)CCCCC3=C2C4=C(N3)C=CC(C#CC5CC5)=C4
InChI:
SGC-PI5P4Kγ/MYLK4-1: InChI=1S/C20H20N4O/c21-20-22-11-14-2-1-3-17-18(19(14)24-20)15-10-13(4-5-16(15)23-17)12-6-8-25-9-7-12/h4-6,10-11,23H,1-3,7-9H2,(H2,21,22,24)
SGC-PI5P4Kγ/MYLK4-1N: InChI=1S/C21H20N4/c22-21-23-12-15-3-1-2-4-18-19(20(15)25-21)16-11-14(8-7-13-5-6-13)9-10-17(16)24-18/h9-13,24H,1-6H2,(H2,22,23,25)
InChIKey:
SGC-PI5P4Kγ/MYLK4-1: VGFHPNRWHOQLEB-UHFFFAOYSA-N
SGC-PI5P4Kγ/MYLK4-1N: ZPQLOIBNJAOMDO-UHFFFAOYSA-N
SGC-PI5P4Kγ/MYLK4-1 was profiled in the DiscoverX scanMAX assay against 403 wild-type kinases at 1 μM. Only 7 kinases showed PoC <10 giving an S10(1 μM) = 0.017. When the PoC <35 fraction was examined, 10 kinases were included (S35(1 μM) = 0.025). Potential off-targets within the S35(1 μM) fraction were tested via biochemical enzymatic or binding assays and/or NanoBRET target engagement assays. Data corresponding with off-target kinase activity is shown in the table below.
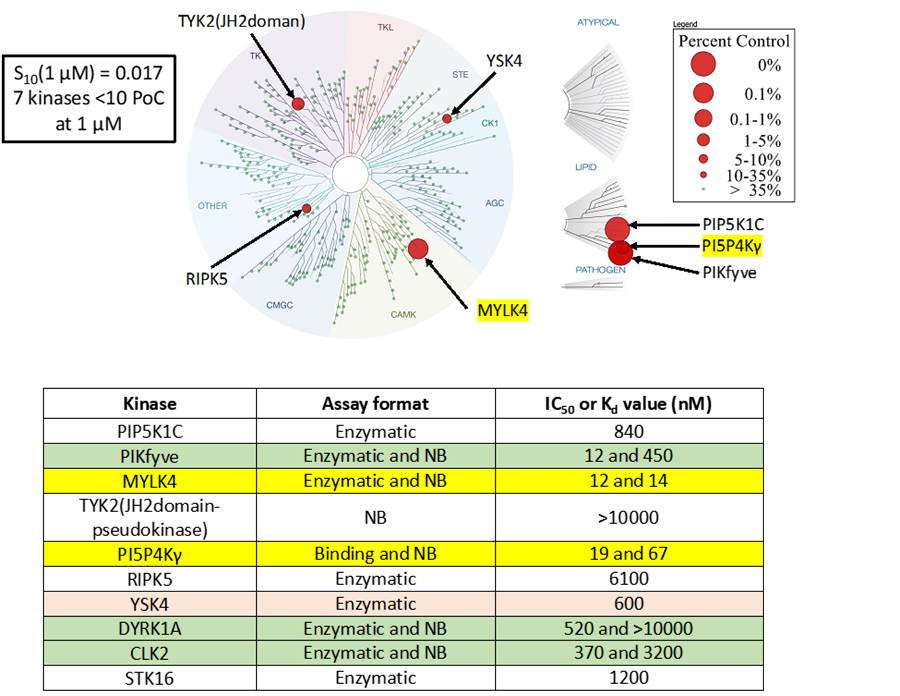
Figure 2: SGC-PI5P4Kγ/MYLK4-1 was profiled in the DiscoverX scanMAX assay against 403 wild-type kinases at 1 μM and off-target kinases with PoC <35 were tested in an orthogonal assay. Rows colored green/yellow are PI5P4Kγ, MYLK4, PIKfyve, CLK2, and DYRK1A. No other kinases demonstrate enzymatic IC50 values within 30-fold of the PI5P4Kγ Kd value. NB = NanoBRET
SGC-PI5P4Kγ/MYLK4-1N was also tested in the DiscoverX scanMAX panel and 5 kinases demonstrated PoC <35 (S35(1 μM) = 0.012). The negative control was sent to SignalChem for testing in the enzyme assay for PIKfyve, to Eurofins DiscoverX or RBC for testing in the enzyme/binding assays for PI5P4Kγ (binding), RIPK5, MEK5, MEK4, and PAK2, and evaluated for MYLK4 affinity via NanoBRET assay. All results are in the table below.
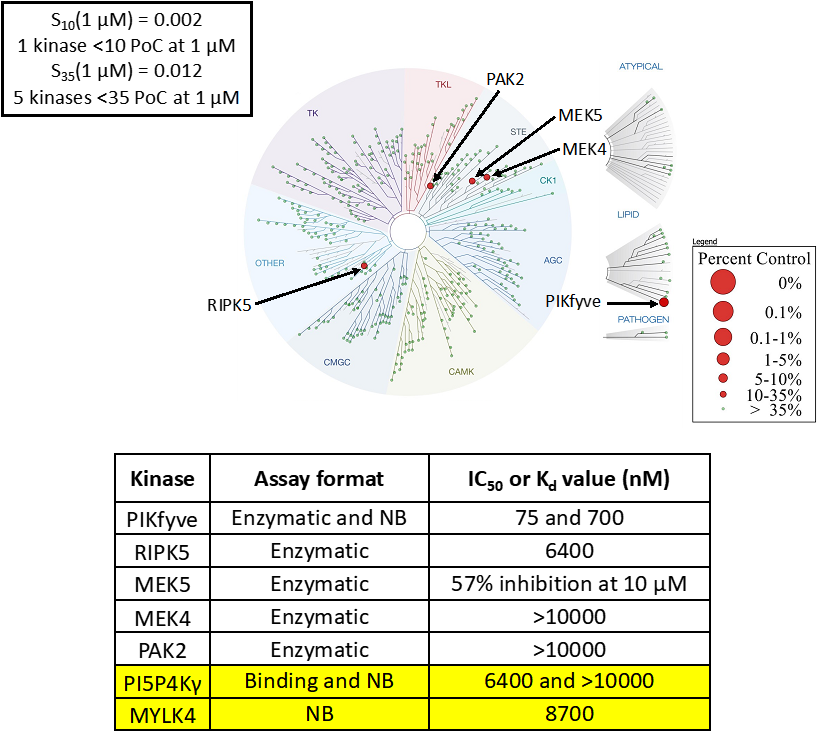
Figure 3: SGC-PI5P4Kγ/MYLK4-1 was profiled in the DiscoverX scanMAX panel against 403 wild-type kinases at 1 μM and follow-up binding/enzymatic assays were done to confirm no activity. NB = NanoBRET
Biological activity summary:
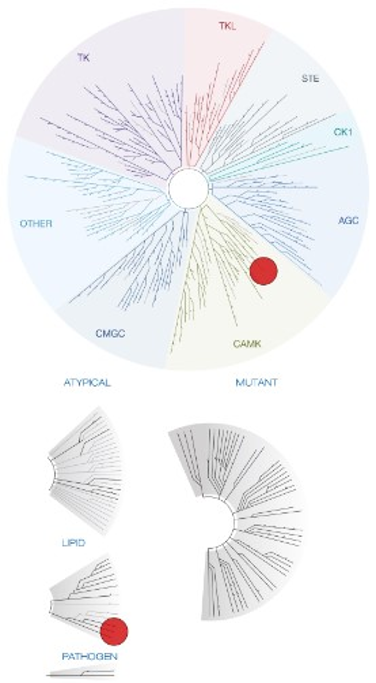
Figure 1: Kinome tree with PI5P4Kγ and MYLK4 highlighted as red circles. Illustration is reproduced courtesy of Eurofins DiscoverX (http://treespot.discoverx.com).
A NanoBRET assay was utilized to assess the binding affinity of SGC-PI5P4Kγ/MYLK4-1 to PI5P4Kγ, MYLK4, PIKfyve, CLK2, DYRK1A, and TYK2 (JH2 domain). The negative control shows no/weak binding affinity for PI5P4Kγ and MYLK4.
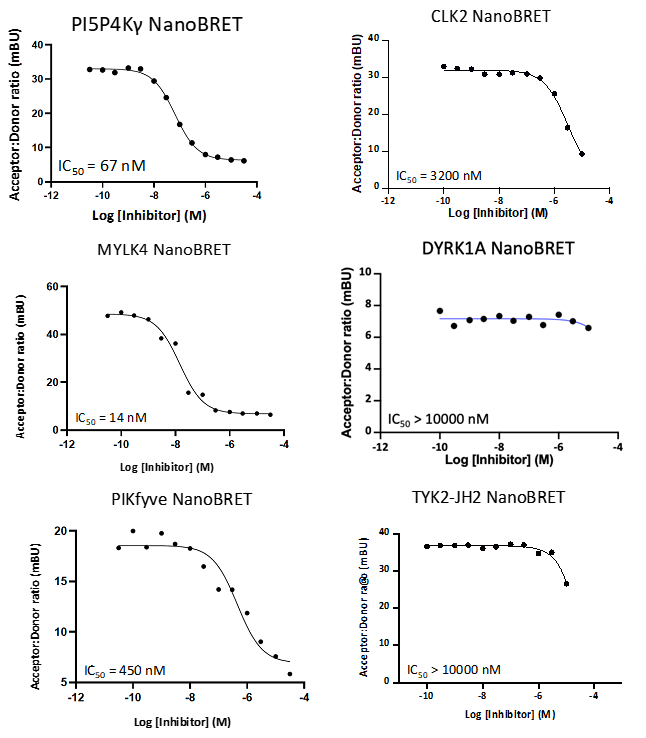
Figure 4: SGC-PI5P4Kγ/MYLK4-1 was profiled in the PI5P4Kγ, MYLK4, PIKfyve, CLK2, DYRK1A, and TYK2 (JH2 domain) NanoBRET assays.
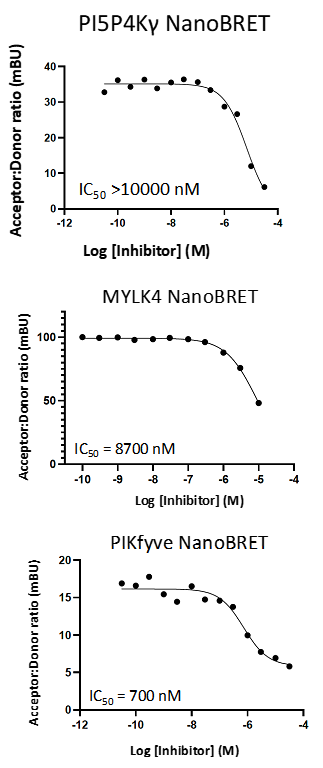
Figure 5: SGC-PI5P4Kγ/MYLK4-1N was profiled in the PI5P4Kγ, MYLK4, and PIKfyve NanoBRET assays.
Drewry, D. H.; Potjewyd, F. M.; Smith, J. L.; Howell, S.; Axtman, A. D. Identification of a chemical probe for lipid kinase phosphatidylinositol-5-phosphate 4-kinase gamma (PI5P4Kγ). Curr Res Chem Biol 2023, 3, 100036; doi: 10.1016/j.crchbi.2022.100036.
Drewry, D. H.; Potjewyd, F. M.; Smith, J. L.; Howell, S.; Axtman, A. D. Identification of a chemical probe for lipid kinase phosphatidylinositol-5-phosphate 4-kinase gamma (PI5P4Kγ). BioRxiv 2022, doi: 10.1101/2022.09.08.507203.