Click here to obtain this probe.
| Probe | Negative control | |
 |
|  |
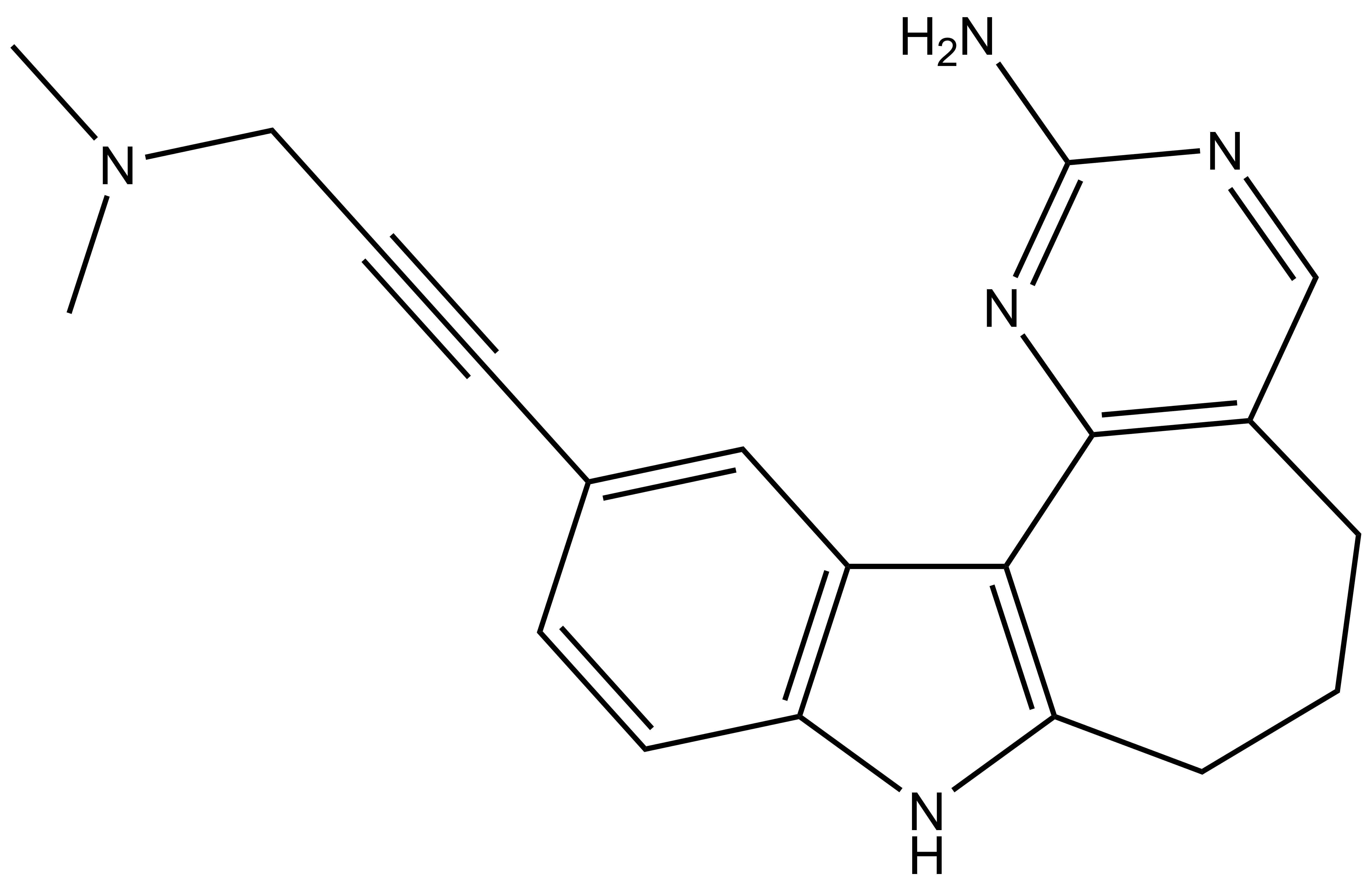
SGC-PIKFYVE-1 |
| SGC-PIKFYVE-1N |
From a library of indolyl pyrimidinamines, we identified a highly potent and cell-active chemical probe (SGC-PIKFYVE-1) that inhibits phosphatidylinositol-3-phosphate 5-kinase (PIKfyve). Comprehensive evaluation of kinome-wide selectivity confirmed that this PIKfyve probe demonstrates excellent selectivity. A structurally similar indolyl pyrimidinamine (SGC-PIKFYVE-1N) was characterized as a negative control that does not inhibit PIKfyve and exhibits exceptional selectivity when profiled broadly. Our PIKfyve chemical probe disrupts multiple phases of the β-coronavirus lifecycle: viral replication and viral entry. Versus published PIKfyve inhibitors, our scaffold is a distinct chemotype that lacks the canonical morpholine hinge-binder of classical lipid kinase inhibitors and has non-overlapping kinase off-targets. Our chemical probe set can be used by the community to further characterize the role of PIKfyve in virology and explore its other roles as well.
 |
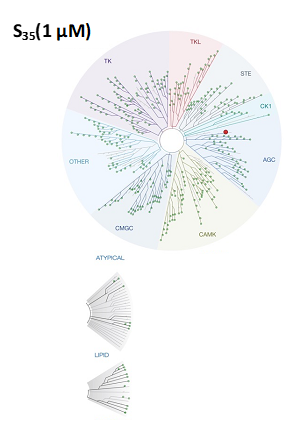
| Figure 1: Kinome tree with PIKfyve highlighted as a red circle. Illustration is reproduced courtesy of Eurofins DiscoverX (http://treespot.discoverx.com) |
Biological activity summary:
• Enzymatic assay (SignalChem): PIKfyve IC50 = 6.9 nM
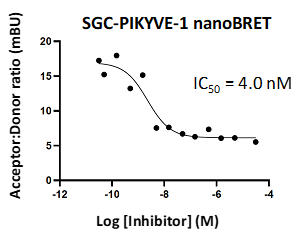
• Cellular data (NanoBRET): PIKfyve IC50 = 4.0 nM
• Only 8/403 kinases with PoC <10 when screened at 1 μM
| Probe |
 |
SGC-PIKFYVE-1 |
| Physical and chemical properties for SGC-PIKFYVE-1 | |
| Molecular weight | 331.4230 |
| Molecular formula | C20H21N5 |
| IUPAC name | 11-(3-(dimethylamino)prop-1-yn-1-yl)-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine |
| MollogP | 1.52 |
| PSA | 66.01 |
| No. of chiral centers | 0 |
| No. of rotatable bonds | 1 |
| No. of hydrogen bond acceptors | 3 |
| No. of hydrogen bond donors | 2 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10 mM) are stable at -20oC |
| Dissolution | Soluble in DMSO up to 10 mM |
| Negative control |
 |
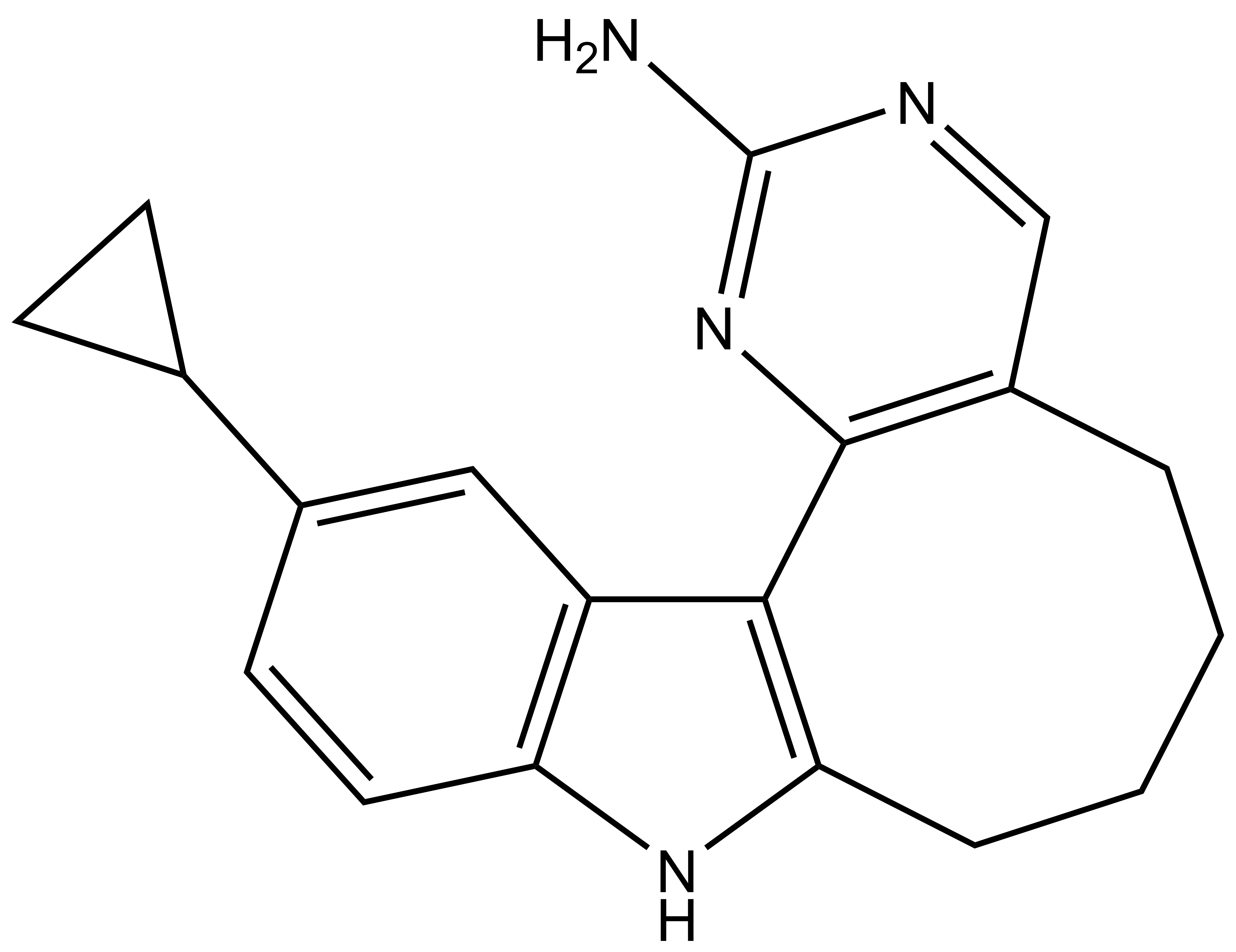
SGC-PIKFYVE-1N |
| Physical and chemical properties for SGC-PIKFYVE-1N | |
| Molecular weight | 304.3970 |
| Molecular formula | C19H20N4 |
| IUPAC name | 12-cyclopropyl-6,7,8,9-tetrahydro-5H-pyrimido[4',5':3,4]cycloocta[1,2-b]indol-2-amine |
| MollogP | 2.47 |
| PSA | 62.77 |
| No. of chiral centers | 0 |
| No. of rotatable bonds | 1 |
| No. of hydrogen bond acceptors | 2 |
| No. of hydrogen bond donors | 2 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10 mM) are stable at -20oC |
| Dissolution | Soluble in DMSO up to 10 mM |
SMILES:
SGC-PIKFYVE-1: CN(C)CC#CC1=CC2=C(C=C1)NC3=C2C4=C(CCC3)C=NC(N)=N4
SGC-PIKFYVE-1N: NC1=NC2=C(C=N1)CCCCC3=C2C4=C(N3)C=CC(C5CC5)=C4
InChI:
SGC-PIKFYVE-1: InChI=1S/C20H21N5/c1-25(2)10-4-5-13-8-9-16-15(11-13)18-17(23-16)7-3-6-14-12-22-20(21)24-19(14)18/h8-9,11-12,23H,3,6-7,10H2,1-2H3,(H2,21,22,24)
SGC-PIKFYVE-1N: InChI=1S/C19H20N4/c20-19-21-10-13-3-1-2-4-16-17(18(13)23-19)14-9-12(11-5-6-11)7-8-15(14)22-16/h7-11,22H,1-6H2,(H2,20,21,23)
InChIKey:
SGC-PIKFYVE-1: DORZPJWJOBMQKC-UHFFFAOYSA-N
SGC-PIKFYVE-1N: XIVFOAKTTGQHLJ-UHFFFAOYSA-N
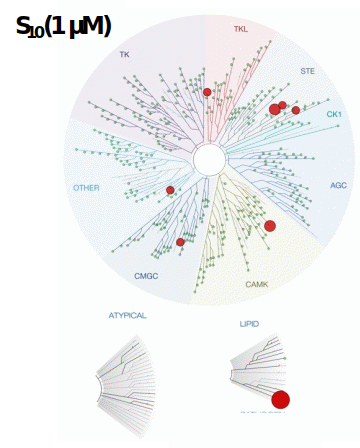
SGC-PIKFYVE-1 was profiled in the KINOMEscan assay against 403 wild-type kinases at 1 μM. Only 8 kinases showed PoC <10 giving an S10(1 μM) = 0.02. When the PoC <35 fraction was examined, 20 kinases were included (S35(1 μM) = 0.05). Potential off-targets within the S35(1 μM) fraction as well as PIKfyve were tested using a NanoBRET target engagement assay and via biochemical enzymatic assays where an assay was available. Data corresponding with off-target kinase activity is shown in the table below.
 |
| Kinase | DiscoverX PoC value | Assay format | IC50 or Kd value (nM) |
| PIKfyve | 0.1 | Enzymatic and NB | 6.9 and 4.0 |
| MYLK4 | 1.8 | Enzymatic and NB | 66 and 270 |
| MEK1 | 4.5 | Enzymatic | >10000 |
| RIPK5 | 5.2 | Enzymatic | >10000 |
| IRAK3 | 9.2 | NT | - |
| MEK2 | 9.6 | Enzymatic | >10000 |
| DYRK1A | 9.9 | Enzymatic | 2040 |
| YSK4 | 9.9 | Enzymatic | 4020 |
| ULK3 | 11 | Enzymatic | >10000 |
| MEK4 | 12 | Enzymatic | >10000 |
| HASPIN | 13 | Enzymatic | 1400 |
| STK16 | 14 | Enzymatic | 560 |
| CLK2 | 20 | Enzymatic | 290 |
| CDK7 | 20 | Enzymatic | >10000 |
| IRAK4 | 21 | Enzymatic | 4500 |
| AURKB | 23 | Enzymatic | 1400 |
| DYRK1B | 26 | Enzymatic | 380 |
| CLK1 | 30 | Enzymatic | 420 |
| RIOK2 | 32 | NT | - |
| CLK4 | 34 | Enzymatic | 440 |
| PIP4K2C | 53 | NB and binding | >10000 and 1900 |
| MAP4K5 | 96 | Enzymatic and NB | 89 and >10000 |
Figure 2: SGC-PIKFYVE-1 was profiled in the KINOMEscan assay against 403 wild-type kinases at 1 μM and off-target kinases inhibited PoC <35 were tested in an orthogonal assay. Rows colored green are PIKfyve and the two kinases with enzymatic IC50 values within 30-fold of the PIKfyve enzymatic IC50 value: MYLK4 and MAP4K5.
SGC-PIKFYVE-1N was also tested in the DiscoverX panel and 1 kinase had a PoC <35 (S35(1 μM) = 0.002). The negative control was sent to SignalChem for testing in enzyme assay for PIKfyve. All results are in the table below.
 |
| Kinase | DiscoverX PoC value | Assay format | IC50 value (nM) |
| MAST1 | 32 | NT | NT |
| PIKfyve | 88 | Enzymatic and NB | 715 and >10000 |
Figure 3: SGC-PIKFYVE-1N was profiled in the KINOMEscan assay against 403 wild-type kinases at 1 μM and a follow-up PIKfyve enzymatic assay was done to confirm no activity.
A NanoBRET assay was utilized to assess the binding affinity of SGC-PIKFYVE-1 to PIKfyve. The negative control shows no binding affinity for PIKfyve.
 |
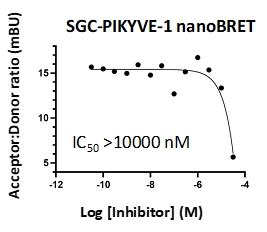 |
Figure 4: SGC-PIKFYVE-1 and SGC-PIKFYVE-1N were profiled in the PIKfyve NanoBRET assay.
Drewry, D. H.; Potjewyd, F. M.; Smith, J. L.; Dickmander, R. J.; Bayati, A.; Howell, S.; Taft-Benz, S.; Min, S. M.; Hossain, M. A.; Heise, M.; McPherson, P. S.; Moorman, N. J.; Axtman, A. D. Identification and utilization of a chemical probe to interrogate the roles of PIKfyve in the lifecycle of β-coronaviruses. J Med Chem 2022, ASAP; doi: 10.1021/acs.jmedchem.2c00697.
Drewry, D. H.; Potjewyd, F. M.; Smith, J. L.; Dickmander, R. J.; Bayati, A.; Howell, S.; Taft-Benz, S. A.; Hossain, M. A.; Heise, M. T.; McPherson, P. S.; Moorman, N. J.; Axtman, A. D. Identification and utilization of a chemical probe to interrogate the roles of PIKfyve in the lifecycle of β-coronaviruses. ChemRxiv 2022. doi: 10.26434/chemrxiv-2022-bj274.