This probe is available from Tocris, Sigma and Cayman Chemical.
This probe (as trifluoroacetate salt) is available from Cayman Chemical.
The control may be requested by clicking here.
| Probe |
 |
SGC0946 |
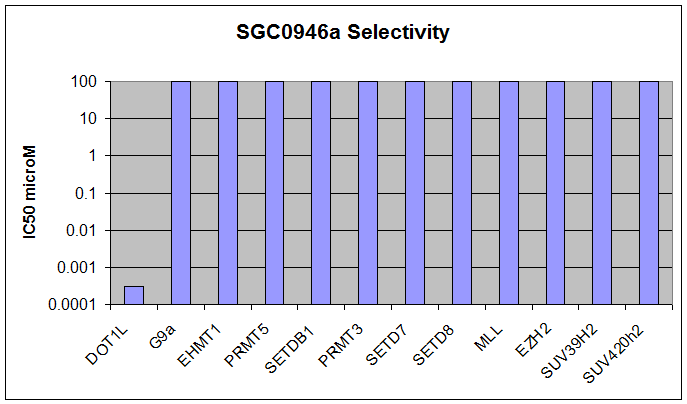
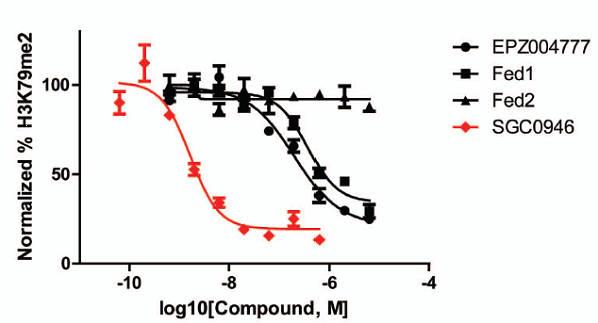
Protein methyltransferases (PMTs) play essential roles in epigenetic regulation of gene expression and chromatin dependent signaling via their methylation of histones and other chromatin associated substrates. Mutation or misregulation of PMTs is linked to many diseases, especially cancer, and there is strong interest in this family of proteins as potential drug targets. Most PKMTs contain a conserved SET-domain, with the exception of DOT1L which has a protein fold that more closely resembles PRMTs. DOT1L is also unique in that it is the only PKMT to methylate histone H3 on Lysine 79 (H3K79), a chromatin mark associated with active chromatin and transcriptional elongation. Here we present SGC0946, a potent and selective inhibitor of DOT1L, which potently and selectively kills cells containing an MLL translocation. SGC0946 inhibits DOT1L with an IC50 of 0.3nM in a radioactive enzyme assay and is over 100-fold selective for other HMTs. In addition, SGC0946 potently reduces H3K79 dimethylation with cellular IC50 of 2.6nM in A431 cells, and 8.8nM in MCF10A cells.
|
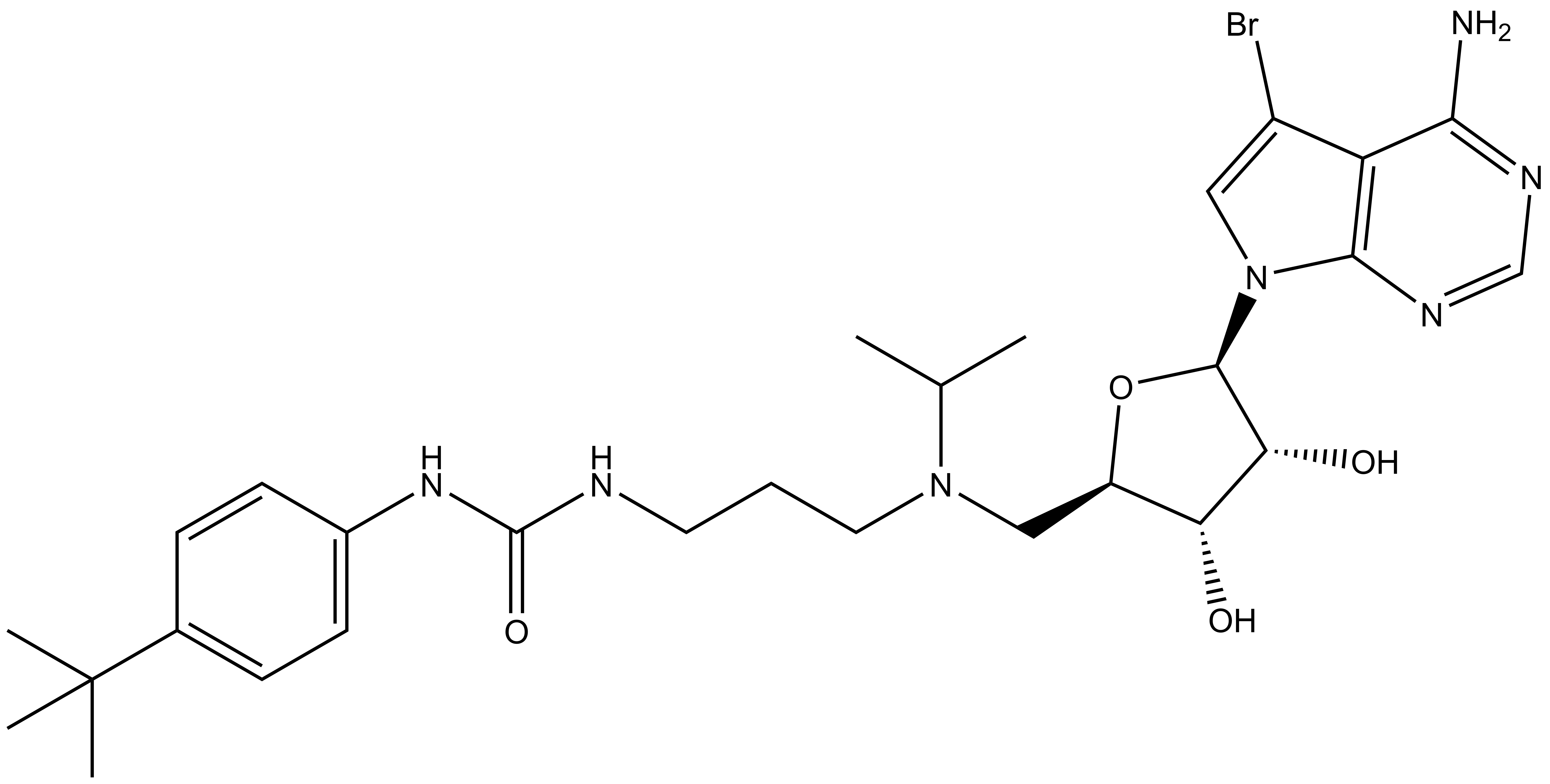
1-(3-((((2R,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(isopropyl)amino)propyl)-3-(4-(tert-butyl)phenyl)urea |
| Physical and chemical properties | |
|---|---|
| Molecular weight | 653.2 |
| Molecular formula | C28H40BrN7O4.HCl |
| IUPAC name | 1-(3-((((2R,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(isopropyl)amino)propyl)-3-(4-(tert-butyl)phenyl)urea |
| logP | 3.51 |
| PSA | 120.4 A |
| Storage | Stable as solid in the dark at -20°C. |
SGC0946 selectively inhibits DOT1L
Inhibition of H3K79me2 by a series of DOT1L inhibitors in A431 cells (4 day treatment).
Catalytic site remodeling of the DOT1L methyltransferase by selective inhibitors
Wenyu Yu, Emma J. Chory, Amy K. Wernimont, Wolfram Tempel, Alex Scopton, Alexander Federation, Jason J. Marineau, Jun Qi, Dalia Barsyte-Lovejoy, Joanna Yi, Richard Marcellus, Roxana E. Iacob, John R. Engen, Erno Wienholds, Fengling Li, Javier Pineda, Guillermina Estiu, Tatiana Shatseva, Taraneh Hajian, Rima Al-awar, John E. Dick, Masoud Vedadi, Peter J. Brown, Cheryl H. Arrowsmith *, James E. Bradner *, Matthieu Schapira *. Nature Communications 3 : 1288doi:10.1038/ncomms2304 (2012)
Main features