This probe is available from Cayman Chemical, Sigma and Tocris (acid).
Its control compound UNC1079 is available from Cayman Chemical
| Probe | Negative control | ||
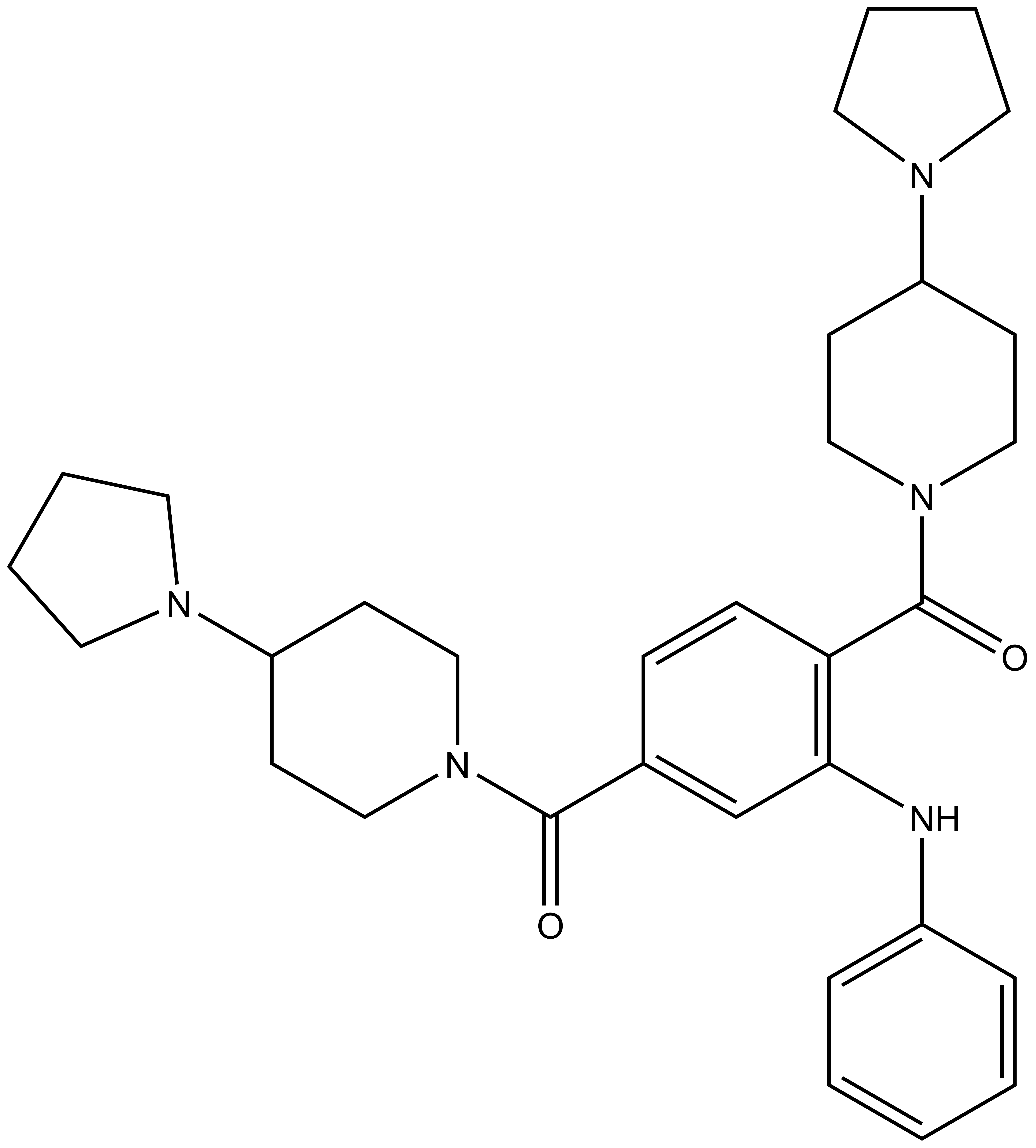 |  | ||
UNC1215 |
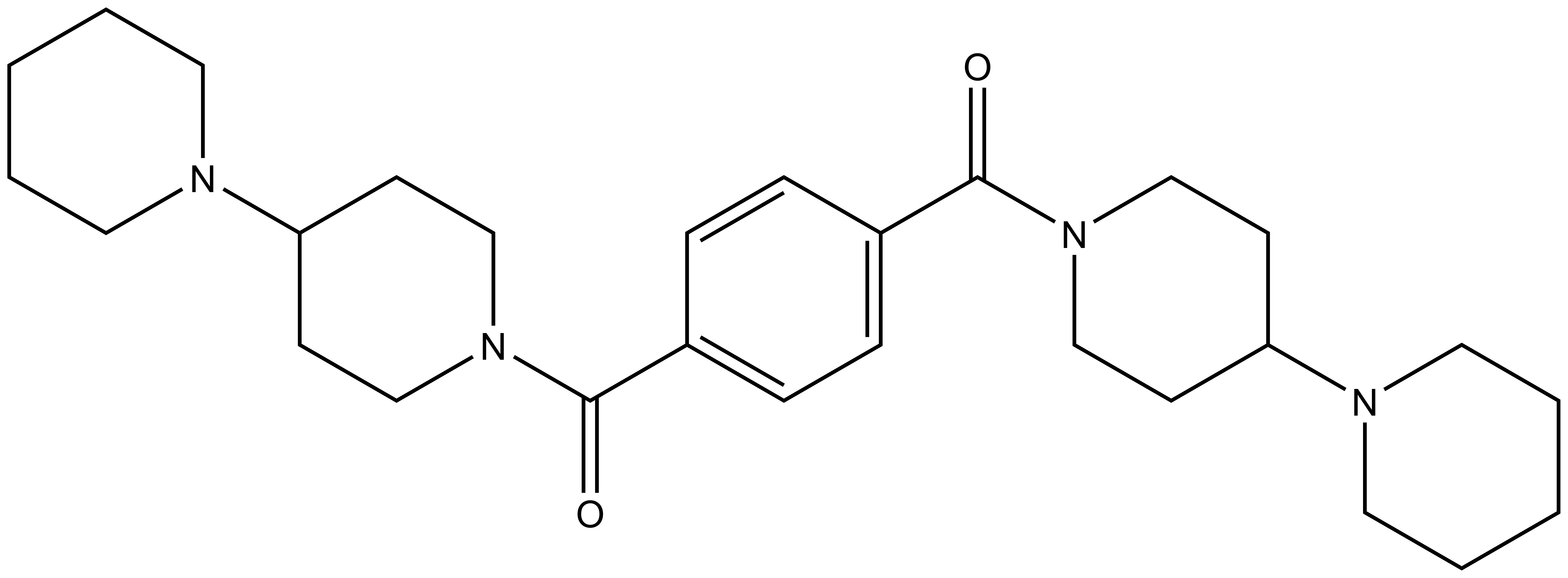
| UNC1079 |
Methyl-lysine (Kme) recognition domains play a central role in epigenetic regulation during cellular differentiation, development, and gene transcription with more than 200 known “reader” domains in the human proteome. In collaboration with the Center for Integrative Chemical Biology and Drug Discovery (CICBDD) at The University of North Carolina at Chapel Hill, we have developed the first chemical probe for a Kme-binding protein. UNC1215 is a potent and selective chemical probe for the Kme reading function of L3MBTL3, a member of the malignant brain tumor (MBT) family of chromatin interacting transcriptional repressors. UNC1215 binds the MBT domains of L3MBTL3 with a Kd of 120 nM, competitively displacing mono- or dimethyl-lysine containing peptides. This probe is greater than 50-fold selective versus other members of the human MBT family and also demonstrates selectivity against more than 200 other Kme reader domains examined. UNC1079 is a structurally similar but substantially less potent antagonist and negative control.
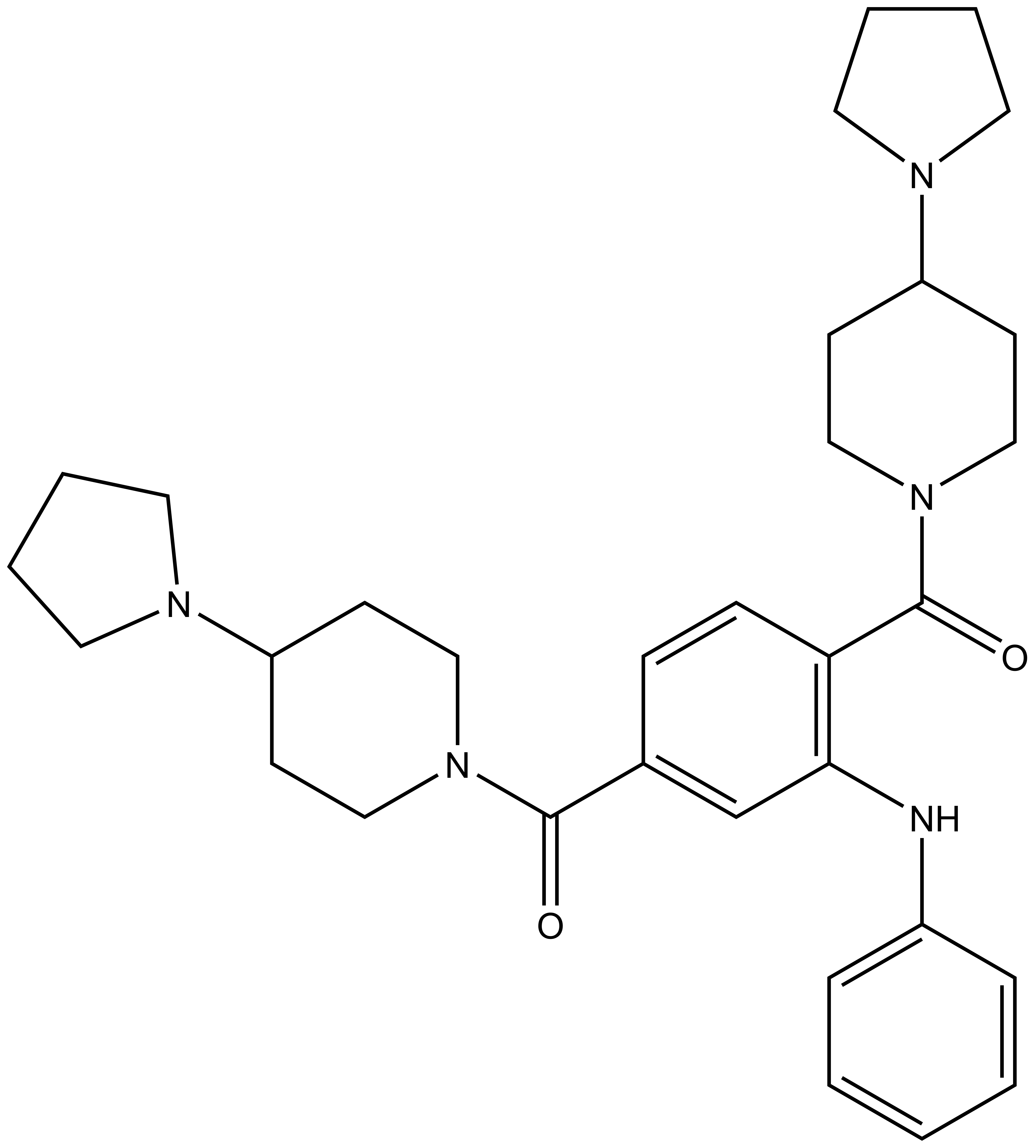
N-phenyl-2,5-bis[4-(pyrrolidin-1-yl)piperidine-1-carbonyl]aniline
Click here to download SDF file
| Physical and chemical properties | |
|---|---|
| Molecular weight | 529.3 |
| Molecular formula | C32H43N5O2 |
| IUPAC Name | N-phenyl-2,5-bis[4-(pyrrolidin-1-yl)piperidine-1-carbonyl]aniline |
| logP | 4.17 |
| PSA | 59.13 |
O=C(N1CCC(N2CCCC2)CC1)C3=C(NC4=CC=CC=C4)C=C(C=C3)C(N(CC5)CCC5N6CCCC6)=O
InChI=1S/C32H43N5O2/c38-31(36-20-12-27(13-21-36)34-16-4-5-17-34)25-10-11-29(30(24-25)33-26-8-2-1-3-9-26)32(39)37-22-14-28(
PQOOIERVZAXHBP-UHFFFAOYSA-N
Selectivity of UNC1215 against a panel of Kme reader proteins.
| Alphascreen IC50 [µM]a | MBT domains | Chromodomain | Tudor Domains | PHD Fingers | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| L3MBTL1 | L3MBTL3 | L3MBTL4 | SFMBT | MBTD1 | CBX7 | 53BP1 | UHRF1 | PHF23 | JARID1A | |
| UNC1215 | 2 | 0.04 | 11 | >30 | 6 | >30 | 4 | >30 | >30 | >30 |
| (R2 = 0.93, n = 17) | (R2 = 0.93, n = 17) | (R2 = 0.63, n = 21) | (n = 18) | (R2 = 0.71, n = 11) | (n = 19) | (R2 = 0.90, n = 12) | (n = 18) | (n = 15) | (n = 15) | |
a - The data for the IC50 values was calculated from n replicate runs; datapoints for each compound concentration were averaged and plotted using 4-parameters curve fitting. R2 is a statistical estimate of goodness-of-fit.
Molecular profiling of UNC1215
| Target Category | Target Activity | ||||
|---|---|---|---|---|---|
Within methyl-lysine reader target family | Target | IC50 (µM)a | Kd (µM)b | ||
| L3MBTL3 | 0.04 | 0.12 | |||
| L3MBTL1 | 2 | 9.4 | |||
| 53BP1 | 4 | > 31 | |||
| PHF20 Tudor | NT | 5.6 | |||
| SPF30 (S.p.) Tudor | NT | Weak binding | |||
| PHF20L1 Tudor | NT | > 13 | |||
| Histone methyltransferases | < 50% inhibition at 250 mM versus 10 targetsc | ||||
| Bromodomains | Tm shift < 0.5 °C at 10 mM versus 12 targetsd | ||||
| Histone demethylases | Tm shift < 0.1 °C at 50 mM versus 15 targetsd | ||||
NIMH Psychoactive Drug Screening Program Selectivity Panel | < 50% inhibition at 10 mM versus 35 targets > 50% inhibition at 10 mM versus 8 targets | ||||
| Target | Ki (µM)e | IC50 (µM) | |||
| Alpha 2C | 0.86 | NT | |||
| DAT | > 10 | NT | |||
| KOR | 4.0 | NT | |||
| M1 | 0.097 | 3.6f | |||
| M2 | 0.072 | 30% at 30 µMg | |||
| M3 | 0.89 | NT | |||
| M4 | 0.40 | NT | |||
| M5 | 4.3 | NT | |||
| Kinase Selectivity Panel | < 15% inhibition at 10 mM versus 49 kinases | ||||
| Target | % Inhibition at 10 mM | ||||
| FLT3 | 64% | ||||
aAlphascreen assays results. bITC results. cRadioactive SAM methyl transfer assay results. dDifferential scanning fluorimetry results. eRadioligand binding assay results. fCa2+ mobilization assay results. gcAMP biosensor assay results.
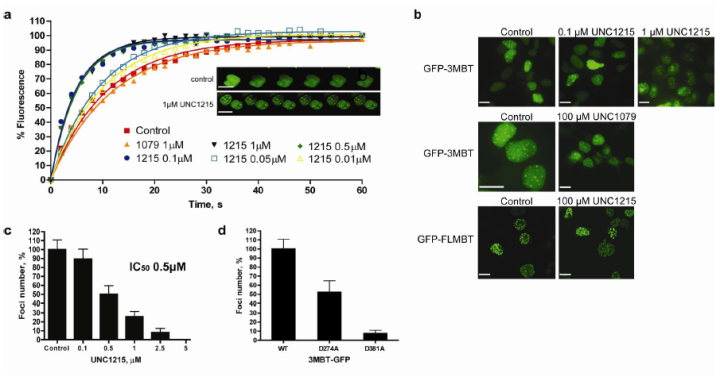
UNC1215 potently antagonizes 3MBT localization in cells. (a) Recovery time of a photobleached area in GFP-3MBT expressing cells is reduced upon treatment with UNC1215 in a dose response manner, whereas inactive compound UNC1079 shows no effect. Inset shows time lapse images of photobleached nuclei. (b, c) GFP fusions of 3MBT and FLMBT localize to the nucleus and form distinct foci in HEK293 cells. UNC1215 inhibits the foci formation of GFP-3MBT in a dose response fashion, whereas UNC1079 has no effect on the foci. In contrast, UNC1215 is relatively ineffective at inhibiting foci formation of GFP-FLMBT (scale bar, 10 µm). (d) The GFP-3MBT D274A MBT1 mutant shows a reduction in foci formation, while the GFP-3MBT D381A MBT2 mutant does not form nuclear foci.
Discovery of a chemical probe for a methyl-lysine reader domain: L3MBTL3
James, Lindsey I.; Barsyte-Lovejoy, Dalia; Zhong, Nan; Krichevsky, Liubov; Korboukh, Victoria K.; Herold, J. Martin; MacNevin, Christopher J.; Norris, Jacqueline L.; Sagum, Cari A.; Tempel, Wolfram; Marcon, Edyta; Guo, Hongbo; Gao, Cen; Huang, Xi-Ping; Duan, Shili; Emili, Andrew; Greenblatt, Jack F.; Kireev, Dmitri B.; Jin, Jian; Janzen, William P.; Brown, Peter J.; Bedford, Mark T.; *Arrowsmith, Cheryl H. *Frye, Stephen V. Nature Chemical Biology 9, 184–191 (2013) - DOI 10.1038/nchembio.1157
Main features