The probe and control may be requested here.
| Probe | Negative control | |
 |
|  |
LH168 |
| LH222 |
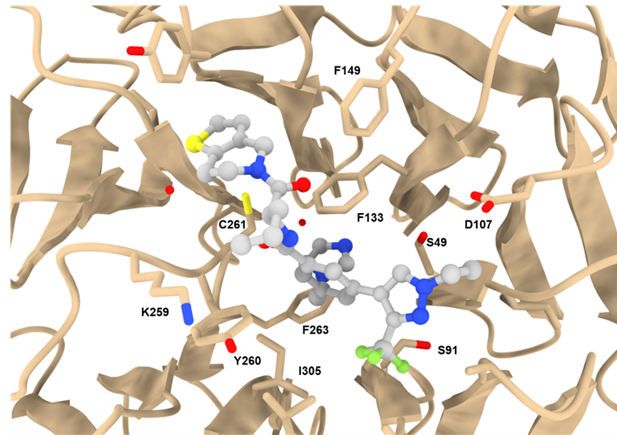
The Histone Lysine Methyltransferase (HMT) complex MLL1 is a crucial epigenetic writer to control DNA accessibility and promote gene transcription [1,2]. The complex incorporates WDR5 (WD40-repeat containing protein 5) that acts as scaffolding component and enhances HMT activity of MLL1. [2] The propeller-shaped WDR5 protein contains two binding sites that interact not only with oncogenic drivers like MLL1, but also with various other proteins like the oncoprotein MYC. [3]
LH168 development started with the identification of a hit from DNA encoded library (DEL)/ML screening [4]. The hit compounds have been subsequently used for further med-chem optimization which provided a potent and selective WDR5 chemical probe LH168 and its negative control compound LH222.
Selectivity for the Homer probe was confirmed by quantitative proteomics.
We recommend that a concentration not higher than 1 µM should be used in cell-based assays.
| Probe | Negative control | |
 |
|  |
LH168 |
| LH222 |
| LH168 | |
| Physical and chemical properties | |
| Molecular weight | 584.665 |
| Molecular formula | C29H31N6O2F3S |
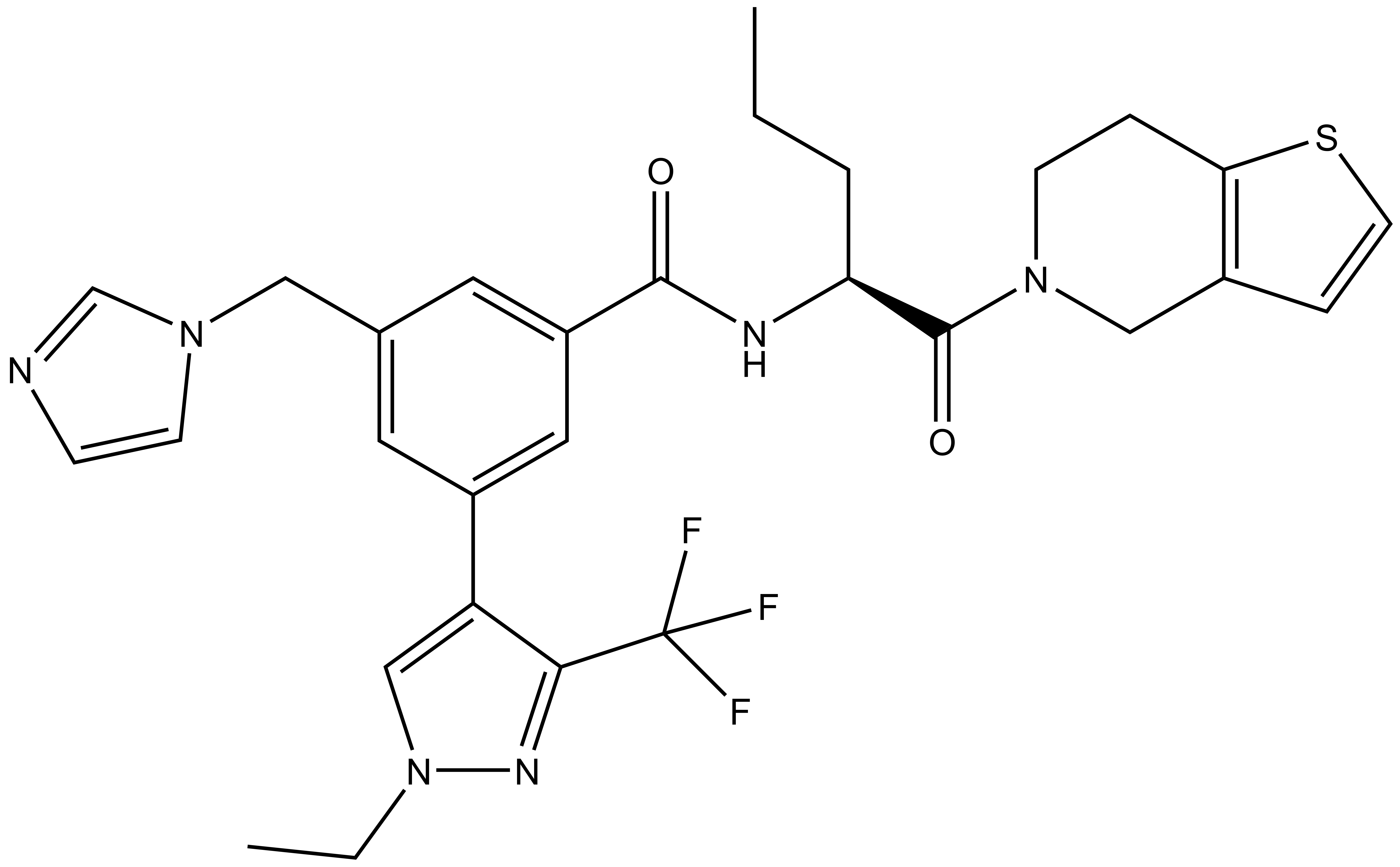
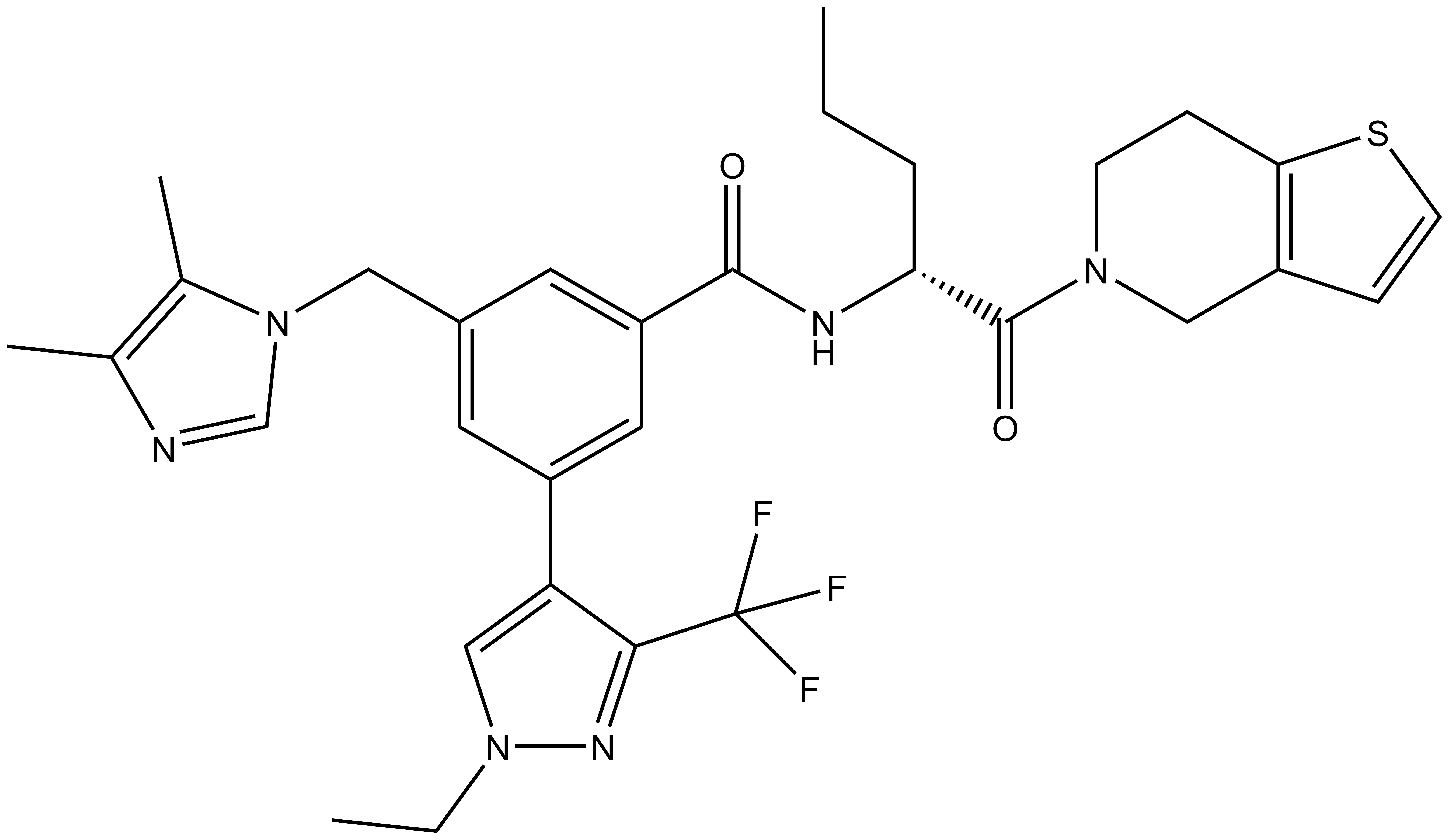
| IUPAC name | (S)-3-((1H-imidazol-1-yl)methyl)-N-(1-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-1-oxopentan-2-yl)-5-(1-ethyl-3-(trifluoromethyl)-1H-pyrazol-4-yl)benzamide |
| clogPo/w | 3.6275 |
| TPSA | 113.29 Å2 |
| No. of chiral centres | 1 |
| No. of rotatable bonds | 10 |
| No. of hydrogen bond acceptors | 8 |
| No. of hydrogen bond donors | 1 |
| Storage | stable as powder at -20°C. |
| Dissolution | soluble in DMSO at 10 mM |
| LH222 | |
| Physical and chemical properties | |
| Molecular weight | 612.719 |
| Molecular formula | C31H35N6O2F3S |
| IUPAC name | (R)-N-(1-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-1-oxopentan-2-yl)-3-((4,5-dimethyl-1H-imidazol-1-yl)methyl)-5-(1-ethyl-3-(trifluoromethyl)-1H-pyrazol-4-yl)benzamide |
| clogPo/w | 4.4233 |
| TPSA | 113.29 Å2 |
| No. of chiral centres | 1 |
| No. of rotatable bonds | 10 |
| No. of hydrogen bond acceptors | 8 |
| No. of hydrogen bond donors | 1 |
| Storage | stable as powder at -20°C. |
| Dissolution | soluble in DMSO at 10 mM |
SMILES:
LH168: O=C([C@H](CCC)NC(C1=CC(CN2C=CN=C2)=CC(C3=CN(CC)N=C3C(F)(F)F)=C1)=O)N4CCC(SC=C5)=C5C4
LH222: O=C([C@@H](CCC)NC(C1=CC(CN2C(C)=C(C)N=C2)=CC(C3=CN(CC)N=C3C(F)(F)F)=C1)=O)N4CCC(SC=C5)=C5C4
InChIKey:
LH168: UFKFTBGGMLIIME-DEOSSOPVSA-N
LH222: RVPFWIYUOKYZMI-AREMUKBSSA-N
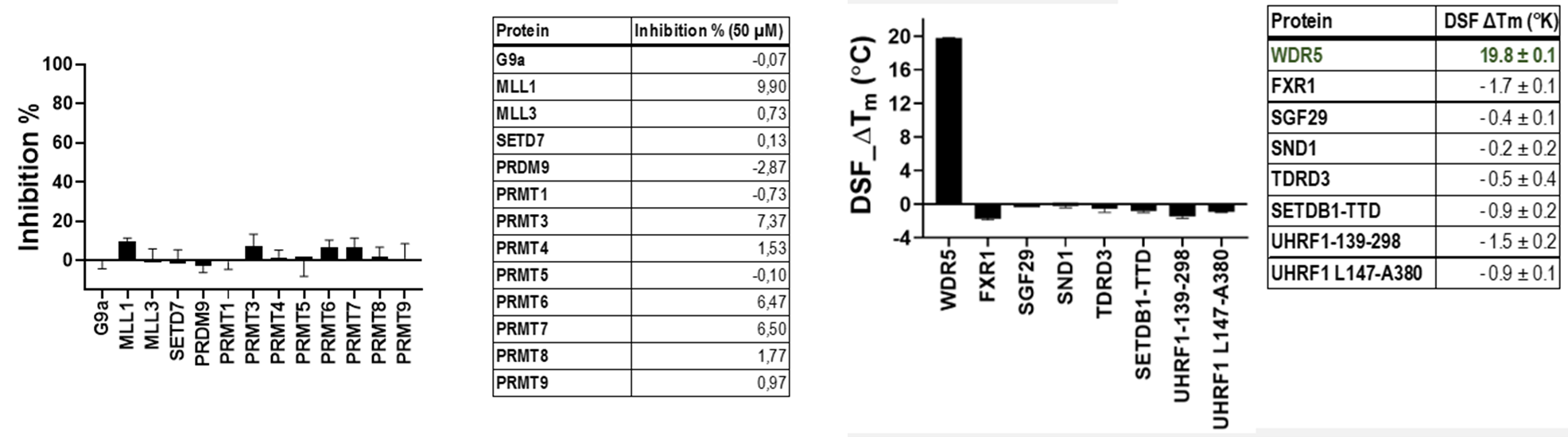
LH168 was tested against 13 histone/arginine methyltransferases by radioactivity-based assay, none of them was significantly inhibited by LH168 at 50 μM. In addition, WDR5 and 7 arginine binders were evaluated in DSF assay using LH168 at 50 μM concentration. As a result, only WDR5 showed significant Tm shift.
Proteomic analysis of a sample enriched by pull down experiment
A cell lysate was subjected to pull down using streptavidin beads and biotinylated analogue of chemical probe. Subsequent proteomic analysis revealed that the samples was enriched for WDR5 as expected.
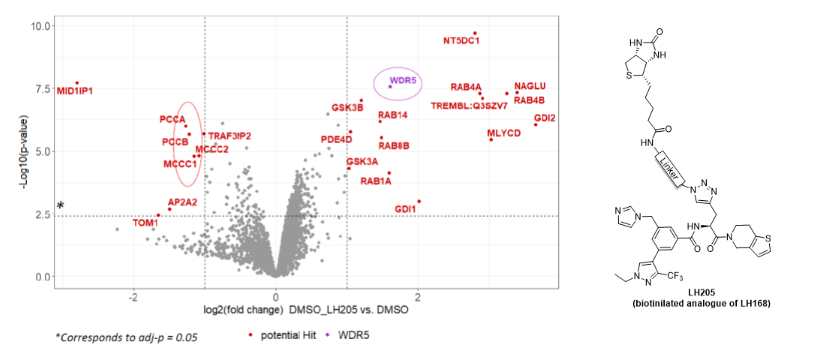
Proteomic analysis of a sample enriched by pull down experiment
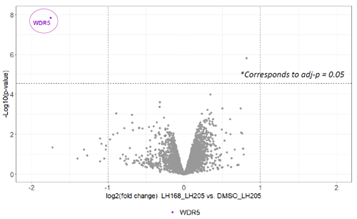
To confirm the selectivity, the pull down experiment was repeated, this time with the co-treatment with excess of LH168 chemical probe which selectively outcompetes the binding to LH205 and therefore prevents the pull down of WDR5 using streptavidin beads.
Assay | Potency |
DSF | 19.8 K |
ITC | Kd = 38 nM |
SPR | Kd = 13 nM |
NanoBRET with intact cells | EC50 = 10 nM |
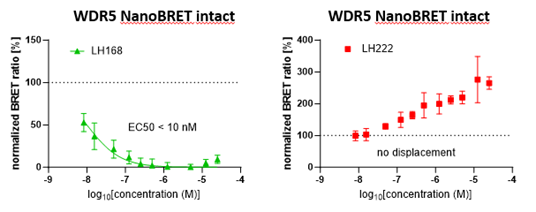
LH168 showed an excellent cellular potency EC50 < 10 nM as measured by NanoBRET target engagement assay. The negative control LH222 showed no tracer displacement.
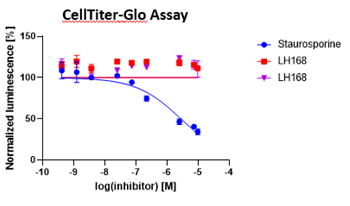
Probe compound LH168 didn’t show any crude cytotoxic effect in HEK293T cells.
[1] Wu, Shu, “MLL1/WDR5 complex in leukemogenesis and epigenetic regulation”, Chin. J. Cancer 2011, 30(4), 240-246.
[2] Jiang, “The complex activities of the SET1/ MLL complex core subunits in development and disease“, BBA – Gene Regulatory Mechanisms 2020, 1863, 194560.
[3] Thomas et al., “Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC”, Mol. Cell 2015, 58, 440-452.
[4] Ackloo et al., "A Target Class Ligandability Evaluation of WD40 Repeat-Containing Proteins", J Med Chem 2025, 68, 1092-1112.
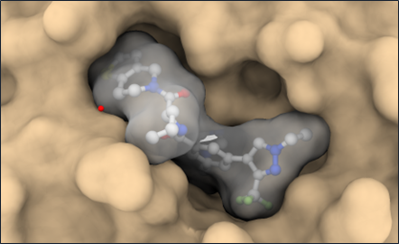
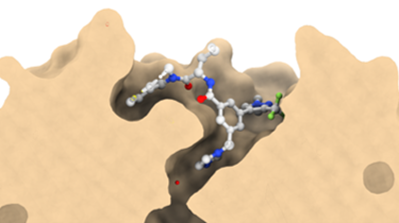
Co-crystal structure with a resolution of 1.6 Å shows the binding mode of LH168 in the WIN-site pocket of WDR5.