Transforming Hit-Finding into a Computational Endeavor: A Roadmap for Open Science Drug Discovery
Finding potent chemical hits for disease-relevant human proteins, especially those that have historically been understudied, remains one of the most persistent challenges in small-molecule drug discovery. Traditional approaches rely on expensive and time-consuming experimental screening of vast chemical libraries, often followed by iterative cycles of synthesis and testing. These methods are largely inaccessible to most academic groups and are inefficient for exploring the long tail of the human proteome.
The Target 2035 initiative, led by the Structural Genomics Consortium (SGC) and supported by a broad coalition of global collaborators, aims to fundamentally transform this paradigm. This bold, open science movement aims to develop a pharmacological tool for every human protein by the year 2035. Launched in 2020, the first phase of Target 2035 focused on identifying high-quality chemical and biological modulators and tested technologies for scalable hit discovery across various protein classes.
Now entering its second phase, Target 2035 proposes a radical shift: to turn hit-finding into a computationally enabled, data-driven endeavor. A new roadmap published in Nature Reviews Chemistry, and co-authored by 78 authors from 30 organizations, lays out how this transformation can occur. Led by Aled Edwards (SGC) and Dafydd Owen (Pfizer), the roadmap distills insights from three years of detailed community planning across academia, biotech, CROs, and industry. At its core lies a simple but powerful idea: by generating large-scale, high-quality, FAIR (Findable, Accessible, Interoperable, Reusable) datasets through open collaboration, we can enable machine learning to drive chemical hit discovery for even the most understudied protein targets.
Why Computational Hit-Finding?
Traditional high-throughput screening is effective for well-studied proteins, but these methods struggle with less-characterized targets that lack biochemical assays or known ligands, which represent the majority of the human proteome. Computational approaches, powered by machine learning (ML) and artificial intelligence (AI), offer a leap forward. However, the effectiveness of ML models relies heavily on the quality of the data used for training. Therefore, the successful implementation of these computational methods depends entirely on access to large amounts of high-quality, diverse, and openly available protein-ligand interaction data.
Building an Open Roadmap
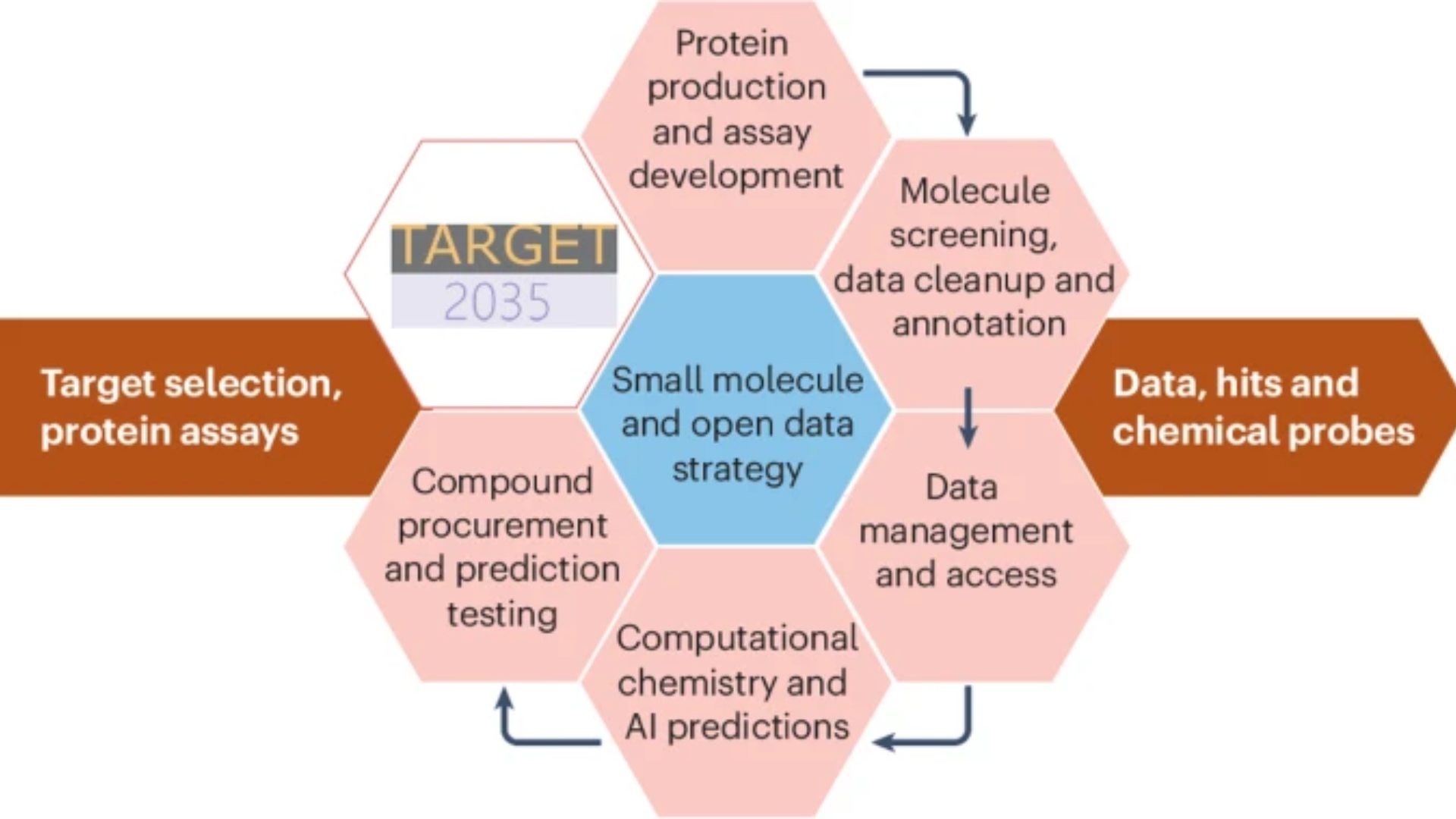
Recognizing the critical need for high-quality data, Target 2035 outlines a five-year plan to generate, structure, and share the data needed to enable ML-based hit-finding. The roadmap integrates best-in-class experimental technologies with open data pipelines and collaborative benchmarking.
Protein Production: More than 2,000 structurally and functionally diverse proteins will be produced and validated by a global network of experimental hubs and community experts. Rigorous quality control, ensuring consistency and reliability across datasets.
Quantitative Binding Data: The Working Group prioritized two direct screening technologies - DNA-Encoded Chemical Libraries (DEL) and Affinity Selection Mass Spectrometry (AS-MS) - to identify binders and generate high-throughput interaction data. These will be complemented by rigorous biophysical studies to characterize binding affinity and mode of action, producing a rich and scalable foundation for ML applications.
Open Data Management: All data, from primary screening to confirmatory testing, will be deposited in AIRCHECK (Artificial Intelligence-Ready CHEmiCal Knowledge base), a purpose-built open-access platform. AIRCHECK will ensure that all datasets are FAIR and available to the global research community for unrestricted reuse and ML model development.
Machine Learning and Benchmarking: The datasets generated will serve as the basis for global ML challenges, modeled after CASP and delivered in partnership with CACHE and DREAM Challenges. Members of the project-associated network of ML scientists, MAINFRAME, will participate in these challenges by training models to predict hits from vast chemical libraries and openly sharing their algorithms. These challenges will include prospective components, where predictions are experimentally tested by consortium labs, closing the iterative loop of prediction, testing, and model refinement.
The Promise of Open Collaboration
What sets this project apart is its commitment to open science, sustained by an international public-private partnership. All participants, whether from academia, biotech, or pharma, agree to share data freely and waive intellectual property claims on the resulting chemical matter. In return, they benefit from accelerated discovery, community tools, and global visibility.
Community participation is welcome at every stage, from protein production and data generation to modeling and validation. This open framework ensures that the benefits of the project extend well beyond the founding partners, creating new opportunities for under-resourced labs, early-career scientists, and innovators worldwide.
A Paradigm Shift for Early Drug Discovery
This roadmap launches Phase 2 of Target 2035, shifting the initiative from concept to execution. Efforts are already underway: protein production is scaling, screening campaigns are active, and curated datasets are being deposited into AIRCHECK. Benchmarking challenges are being designed in partnership with ML and experimental collaborators to ensure rapid feedback loops and iterative improvements.
In the short term, this work will yield validated chemical hits for hundreds of understudied proteins. In the long term, it will deliver powerful computational tools and democratize access to early-stage drug discovery. Such a transformation could rapidly advance the creation of targeted therapeutics, impacting both basic research and clinical outcomes on a global scale.
Read the full publication: https://www.nature.com/articles/s41570-025-00737-z
