This probe is available from Tocris, Sigma and Cayman Chemical
| Probe |
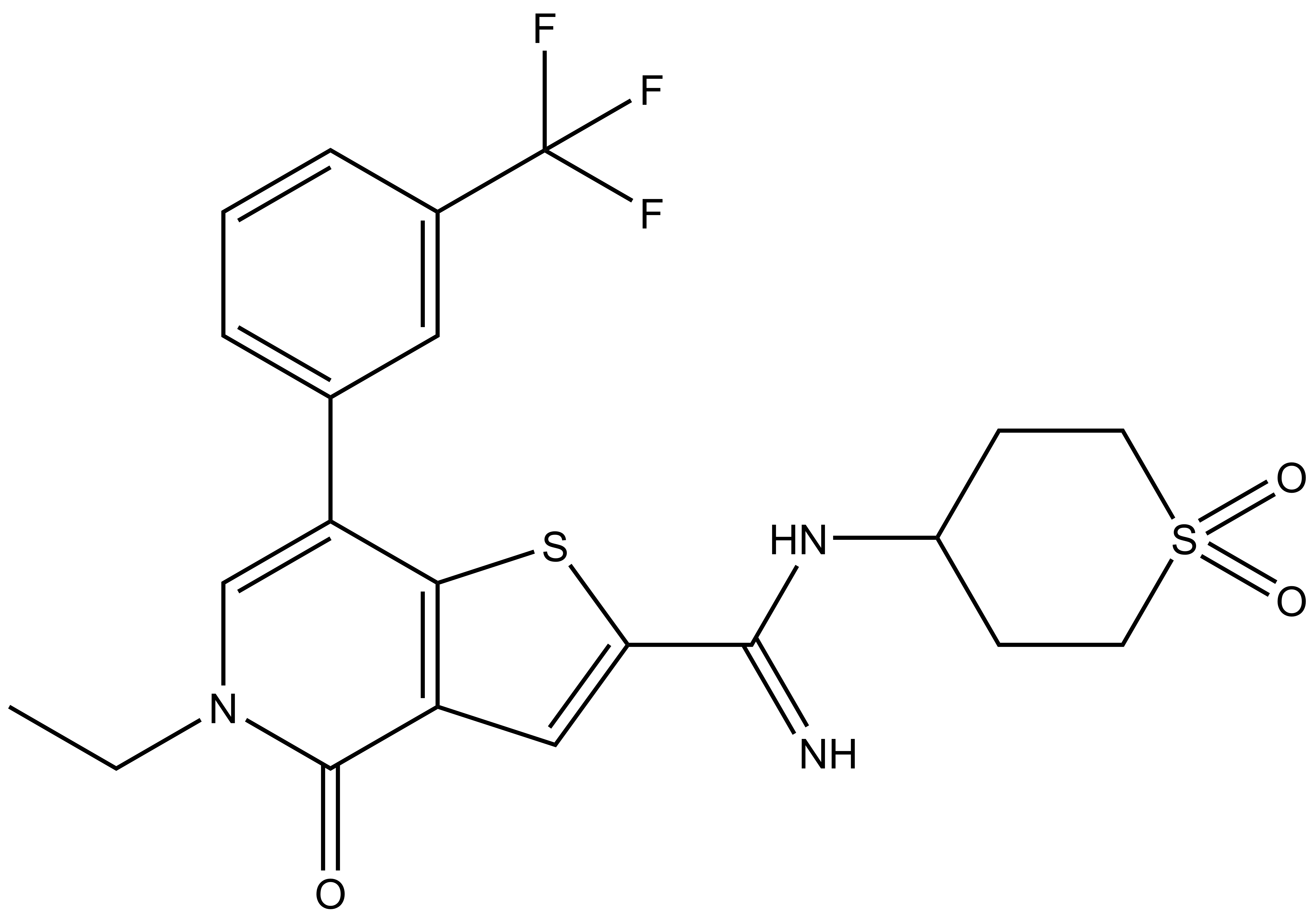 |
I-BRD9 |
Biology of the BRD9 Bromodomains
BRD9 is a bromodomain containing protein that form a small sub-branch of the bromodomain family tree [1]. Human BRD9 contains a single bromodomain and has five isoforms that are produced by alternative splicing. Little is known about BRD9 function but it has been implicated in chromatin remodelling as part of the SWI/SNF complex.
I-BRD9: A Chemical Probe for BRD9
I-BRD9 is a BRD9 specific inhibitor, identified by workers at GlaxoSmithKline [2].
Co-crystal structure
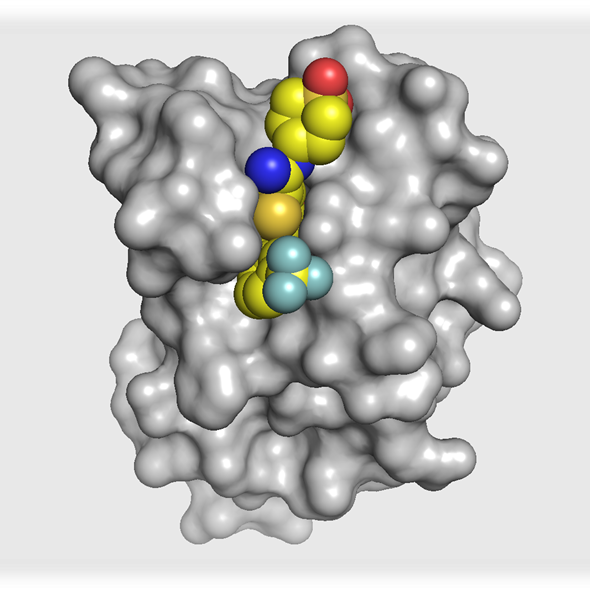
The co-crystal structure of I-BRD9 (magenta) with BRD9 BRD (pdb id 4UIW) has been solved, click on the 'Co-Crystal structures' tab above for more details.
Potency Against Target Family
I-BRD9 is a potent and selective binder to BRD9 with a pIC50 in a Time-Resolved FRET assay of 7.3. BRD4 was used as a representative member of the BET family for initial selectivity screening, I-BRD9 displayed a pIC50 of 5.3 against this protein.
Cellular Activity
I-BRD9 binds to endogenous BRD9 in a chemoproteomic assay and shows good selectivity over the BET family member BRD3.
| I-BRD9 |
 |
N-(1,1-dioxidotetrahydro-2H-thiopyran-4-yl)-5-ethyl-4-oxo-7-(3-(trifluoromethyl)phenyl)-4,5-dihydrothieno[3,2-c]pyridine-2-carboximidamide |
| Physical and chemical properties | |
|---|---|
| Molecular weight | 497.55 |
| Molecular formula | C22H22F3N3O3S2 |
| IUPAC name | N-(1,1-dioxidotetrahydro-2H-thiopyran-4-yl)-5-ethyl-4-oxo-7-(3-(trifluoromethyl)phenyl)-4,5-dihydrothieno[3,2-c]pyridine-2-carboximidamide |
| ChromlogDpH7.4 | 3.7 |
| PSA | |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 5 |
| No. of hydrogen bond acceptors | 10 |
| No. of hydrogen bond donors | 2 |
| Storage | Stable as solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Solubility | Soluble in water to 300uM |
A suitable negative control compound for I-BRD9 has not yet been identified.
Potency against Target
I-BRD9 is a potent and selective binder to (full length) BRD9 with a pIC50 in a Time-Resolved FRET assay of 7.3. BRD4 was used as a representative member of the BET family for initial selectivity screening, I-BRD9 displayed a pIC50 of 5.3 against this protein.

Selectivity
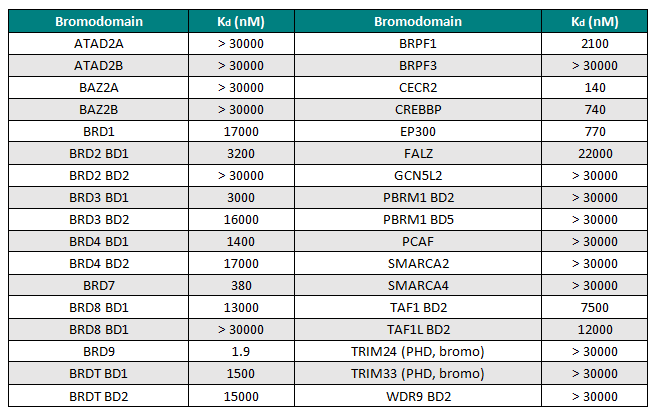
I-BRD9 was profiled by BROMOscan™, a novel bromodomain inhibitor binding platform that measures the interactions between test compounds and a panel of bromodomain assays (http://www.discoverx.com), against 34 bromodomains. The results from this screen indicated nanomolar affinity binding at BRD9, with pKd = 8.7. I-BRD9 showed >700-fold selectivity over the BET family of bromodomains and 200-fold over the highly homologous BRD7 bromodomain (pKd = 6.4). I-BRD9 displayed >70-fold selectivity against every other bromodomain tested.
Selectivity Beyond Target Family
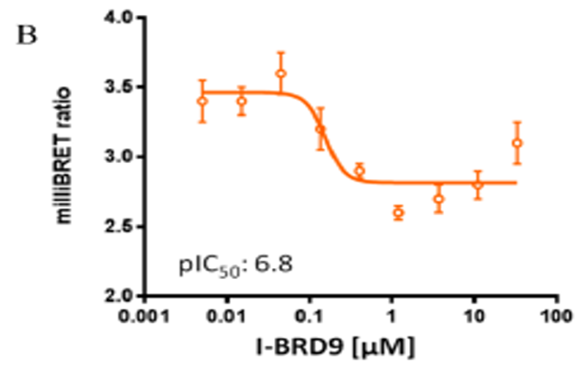
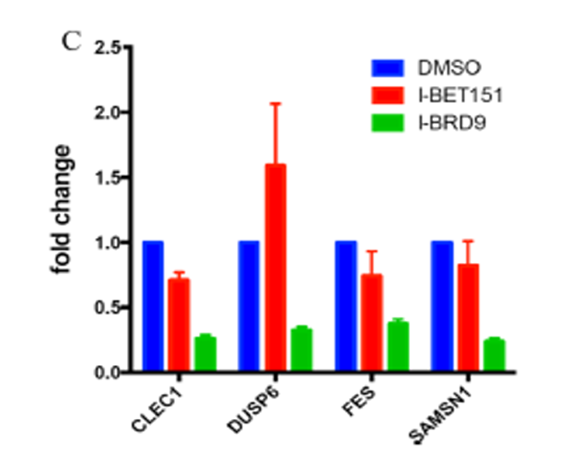
I-BRD9 was found to be inactive against a panel of 49 human receptors, ion channels, kinases and other enzymes. Cellular target engagement of BRD9 and disruption of chromatin binding was demonstrated through a NanoBRET assay; and CLEC1, DUSP6, FES, and SAMSN1 genes are selectively regulated in the presence of I-BRD9
Engagement of BRD9 in a cell assay
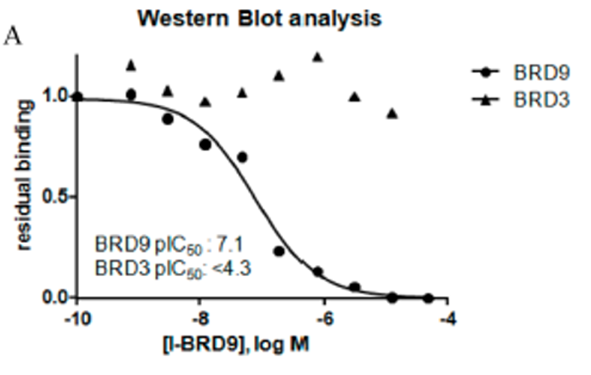
In a chemoproteomic competition binding assay in HUT-78 cell lysate followed by Western blot analysis, the binding of I-BRD9 to endogenous BRD9 displayed >625-fold selectivity against BET family member BRD3. These data confirm potency at BRD9, and selectivity over the BET family is maintained with endogenous proteins.
Cellular target engagement of BRD9 and disruption of chromatin binding was demonstrated through a NanoBRET assay measuring displacement of NanoLuc-tagged BRD9 bromodomain from Halotagged histone H3.3
qPCR validation of CLEC1, DUSP6, FES, and SAMSN1 genes selectively regulated by compound I-BRD9 (10 μM), but not by I-BET151 (1 μM). Genes were previously identified by full gene transcriptomics in Kasumi-1 cells
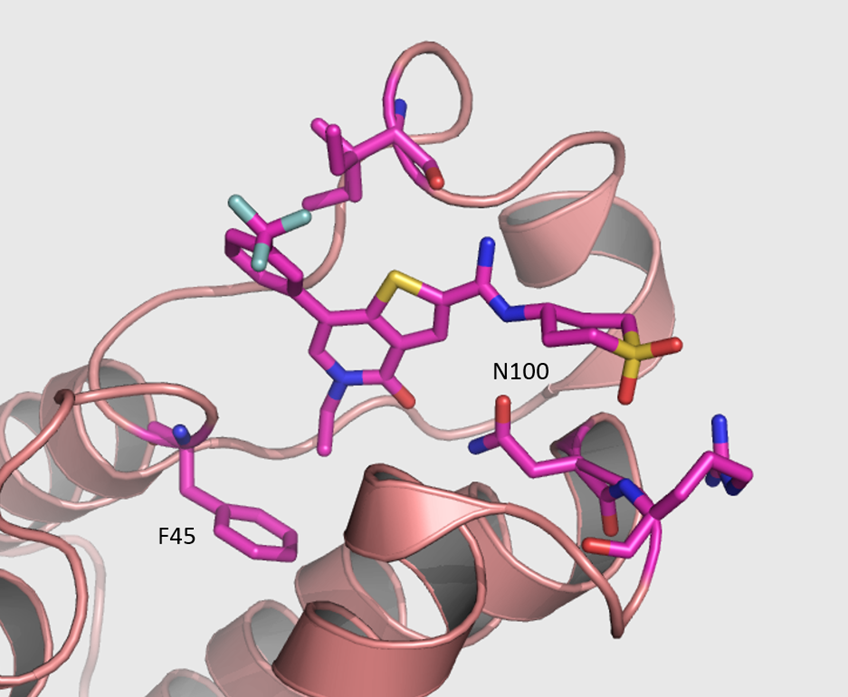
The co-crystal structure of I-BRD9 with BRD9 has been solved with a resolution of 1.73 Å (PDB id: 4UIW). The key features are:
- I-BRD9 makes H-bond interactions to the Asn100 side chain, the backbone carbonyl of Ile53, and the backbone NH of Arg101.
- The N-ethyl thienopyridinone Kac mimetic is comfortably accommodated in the hydrophobic binding pocket with a slight movement of Phe45, having little effect on the conformation of the remainder of the site.
- Other interactions are illustrated in the diagram below
TR-FRET BRD9 Binding Assay BRD9 TR-FRET Assay
A proprietary bromodomain binding small molecule containing a pendant primary amine was tagged with Alexa Fluor 647 (GSK2833930A). All assay components were dissolved in buffer (50 mM HEPES pH7.4, 50 mM NaCl, 5% Glycerol, 1 mM DTT and 1 mM CHAPS). The final concentration of BRD9 protein was 10 nM and the Alexa Fluor647 ligand was at Kd (~100 nM for BRD9). 5 uL of this reaction mixture was added to all wells containing 50 nl of various concentrations of test compound or DMSO vehicle (0.5% DMSO final) in 384 well microtitre plates and incubated in dark for 30 min at room temperature. Eu-W1024 Anti-6xHis Antibody (AD0111 PerkinElmer) at 1.5 nM FAC was used as a detection reagent. The plates were read on the Envision reader and the donor and acceptor counts were determined and the ratio of acceptor/donor was calculated.
Chemoproteomic Profiling
Nuclear extract was produced from fresh HuT78 cells. Affinity profiling assays were performed by derivatising sepharose beads with 2.0 mM of a proprietary bromodomain binding small molecule containing a pendant primary amine (GSK2893910A). I-BRD9 was spiked into HuT78 mixed nuclear and chromatin extracts and incubated for 45 minutes at 4 °C. Derivatised sepharose beads (35 μl beads per sample) were equilibrated in lysis buffer and incubated with cell extract pre-incubated with compound. Beads were washed with lysis buffer containing 0.2 % NP-40 and eluted with 2x SDS sample buffer supplemented with DTT. Aliquots of the eluates from chemoproteomic assays were separated on 4–12 % gel (NuPAGE, Invitrogen) and this was used for Western Blot analysis with antI-BRD9 (Abcam, ab-66443) and anti-BRD3 (Santa Cruz, SC-81202) antibodies.
NanoBRET Assay
HEK293 cells were plated in each well of a 6-well plate and co-transfected with Histone H3.3-HaloTag (NM_002107) and NanoLuc-BRD9 (Q9H8M2) BD domain amino acids 120-240. 20 hrs post-transfection the cells were collected, washed with PBS, and exchanged into media containing phenol red-free DMEM and 4% FBS in the absence (control sample) or the presence (experimental sample) of 100nM NanoBRET 618 fluorescent ligand (Promega). Cell density was adjusted, re-plated in a 96-well plate and inhibitor added directly to media at final concentrations between 0-33 μM and the plates were incubated for 18 hours at 37 °C in the presence of 5% CO2. NanoBRET furimazine substrate (Promega) was added to both control and experimental samples at a final concentration of 10 μM and readings were performed within 5 minutes using the CLARIOstar (BMG) equipped with 450/80 nm bandpass and 610 nm longpass filters with a 0.5sec reading setting. A corrected BRET ratio was calculated and is defined as the ratio of the emission at 610 nm/450 nm for experimental samples (i.e. those treated with NanoBRET fluorescent ligand) subtracted by and the emission at 610 nm/450 nm for control samples (not treated with NanoBRET fluorescent ligand).