This probe is available from Cayman Chemical and Tocris.
Its negative control (ent-LP99) is available for purchase from Tocris.
| Probe | Negative control | |
 |
|  |
LP99 |
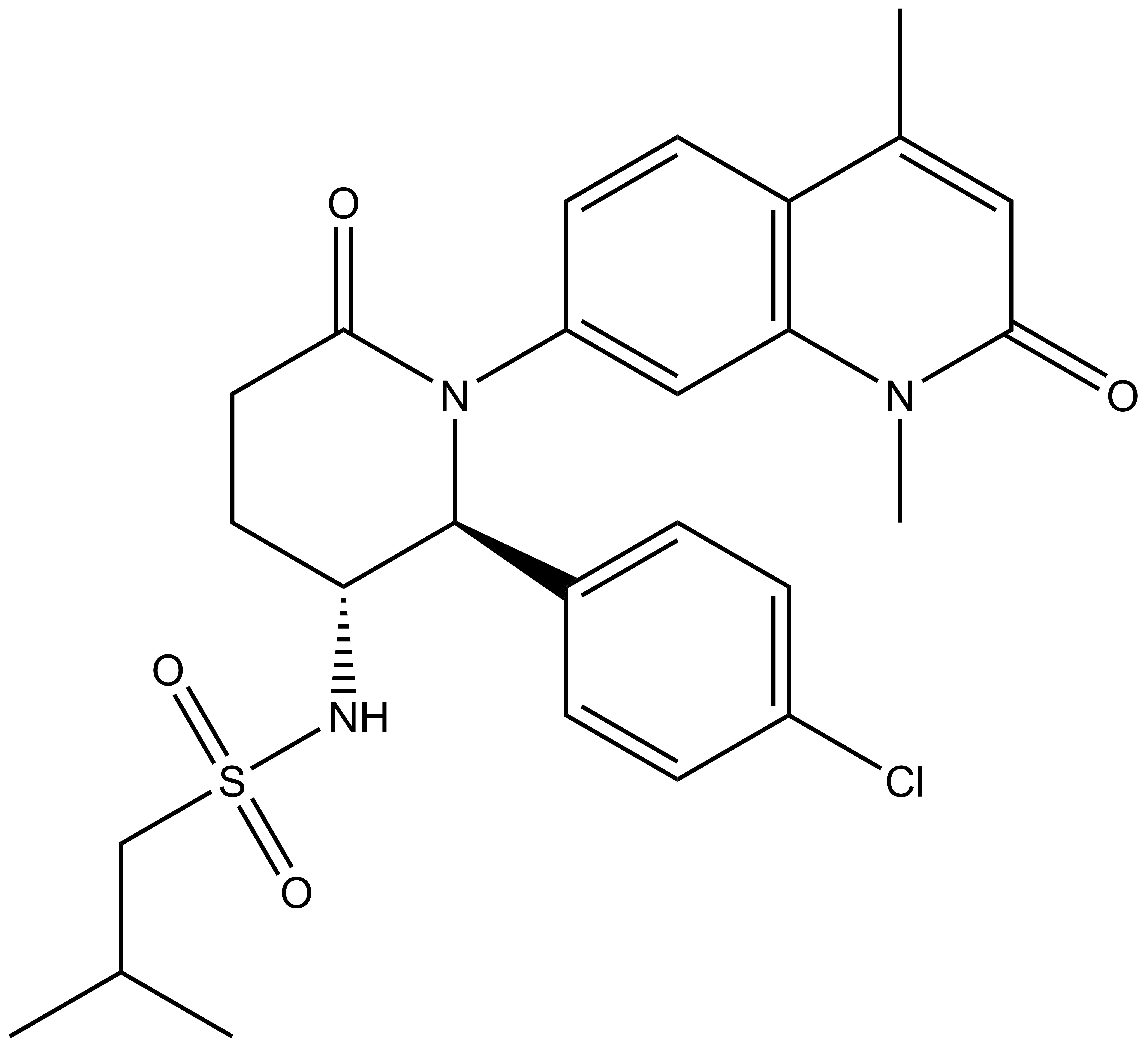
| 2S,3R-Enantiomer |
BRD7 and BRD9 are related bromodomain- containing proteins that form a small sub-branch of the bromodomain family tree [1]. Human BRD9 contains a single bromodomain and has five isoforms that are produced by alternative splicing. Little is known about BRD9 function; BRD7 has been reported to act both as coactivator, for example for some p53 target genes and as corepressor, negatively regulating the GSK3B phosphotransferase activity. Both proteins have been implicated in chromatin remodelling as part of the SWI/SNF complex. BRD7 has been described as transcriptional corepressor that down-regulates the expression of target genes. Its binding to promoters also leads to increased histone H3 acetylation at 'Lys-9' (H3K9ac).
LP99, a BRD7/9 specific inhibitor, is a result of collaboration with Professor Darren J. Dixon at the University of Oxford [2]
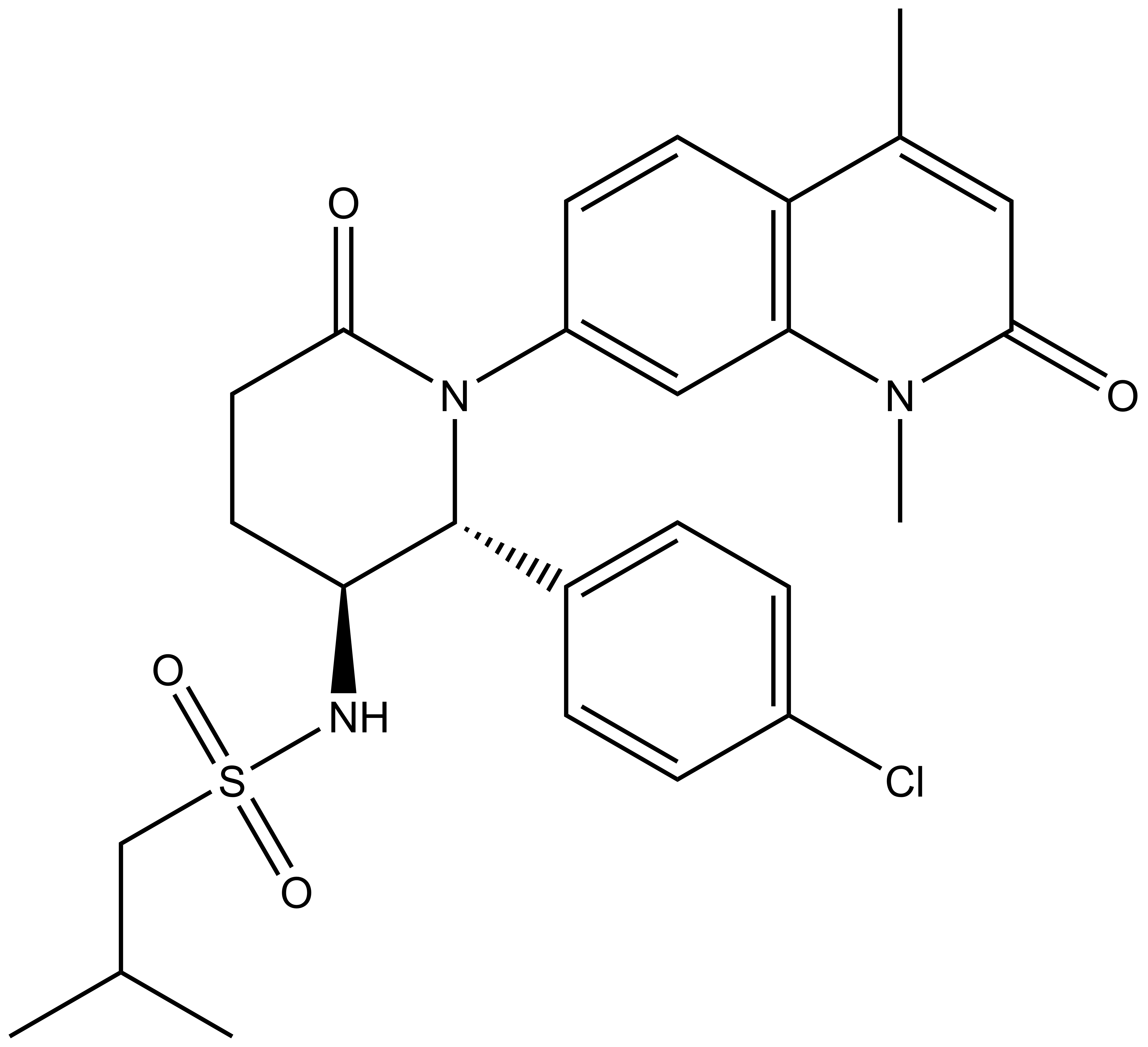
Chemical structure of the active enantiomer (2R, 3S)-LP99. The enantiomer (2S, 3R)-LP99 is inactive against BRD9. The chiral centres are highlighted by a *.
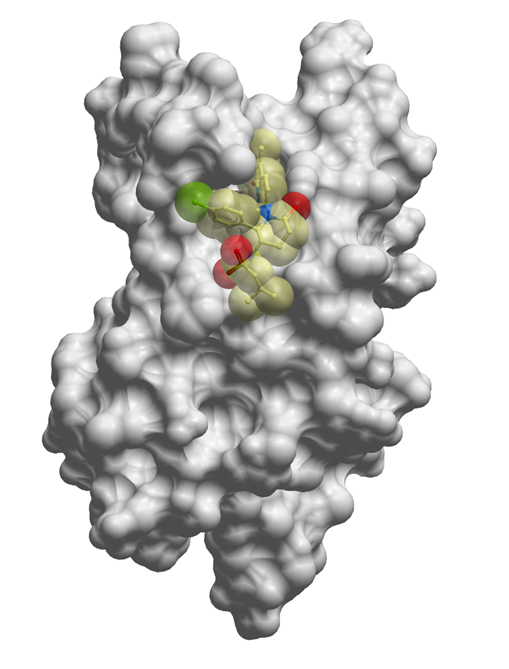
The co-crystal structure of LP99 (yellow) with BRD9 BRD has been solved, click on the 'Co-Crystal structures' tab above for more details.
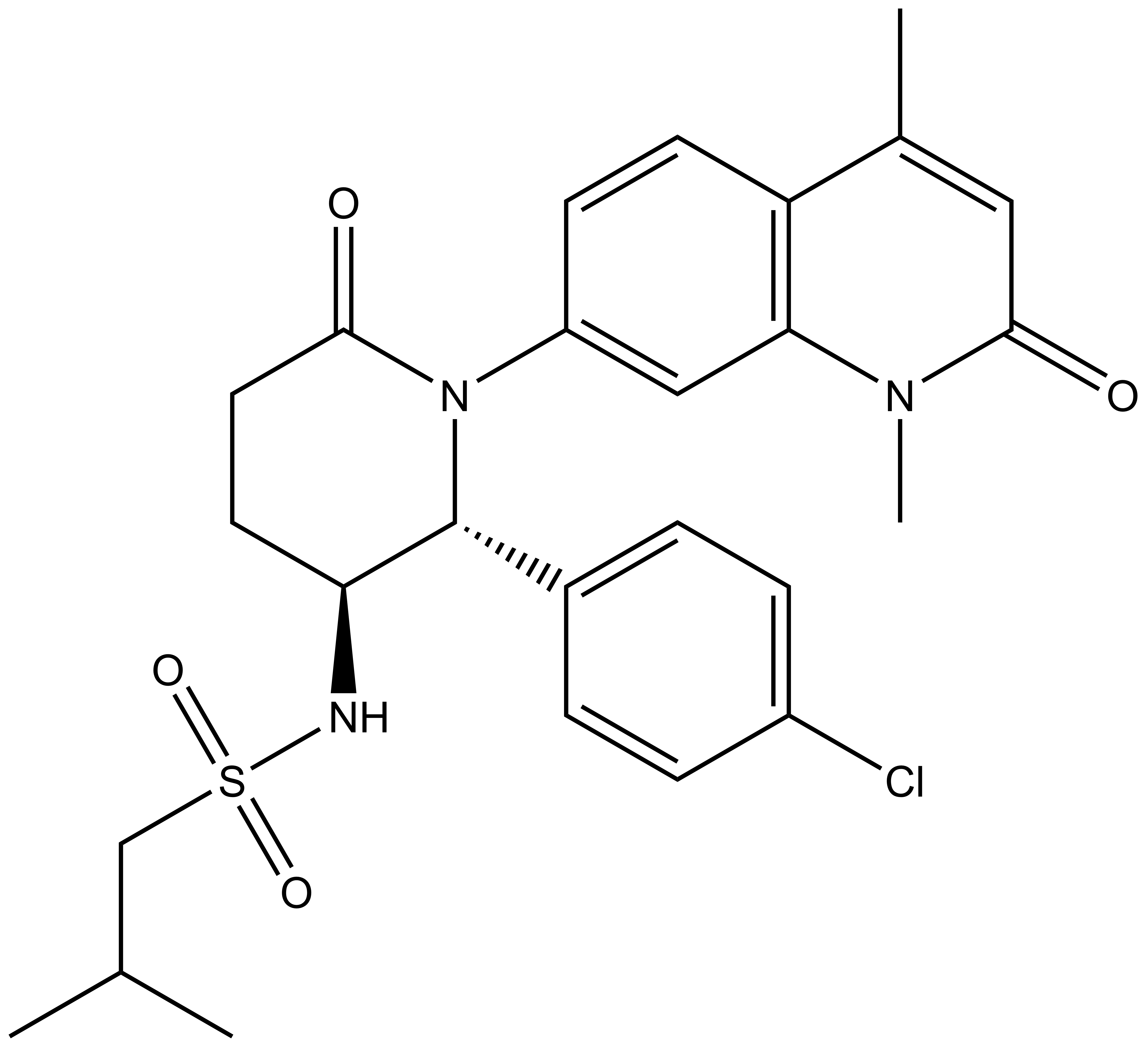
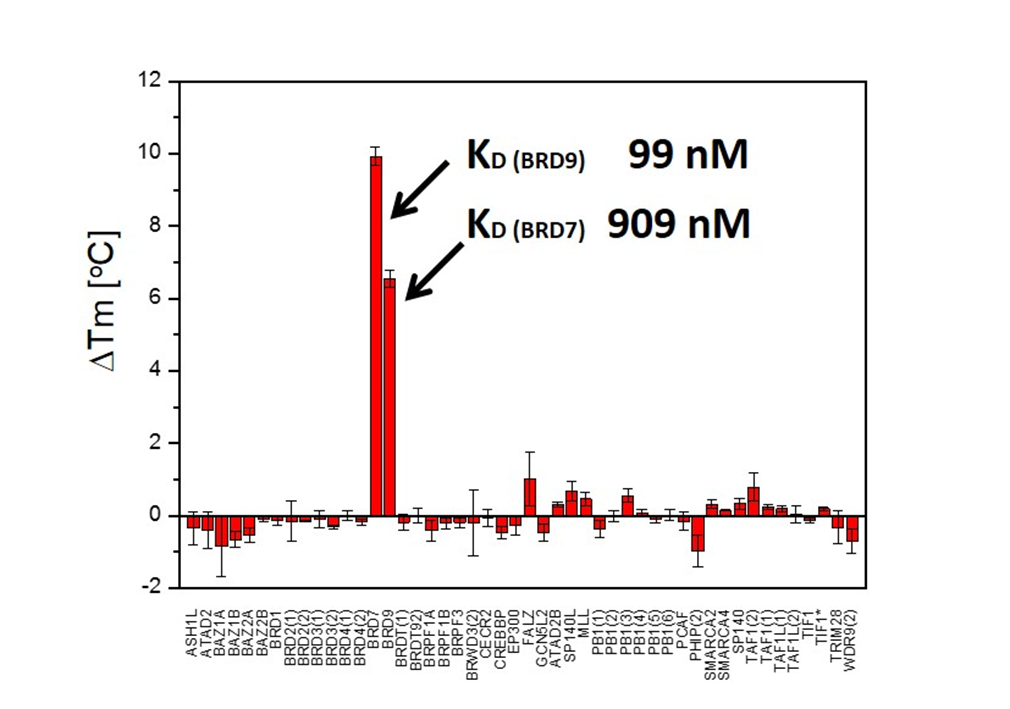
LP99 is a potent binder to BRD9 with a KD = 99 nM as shown by ITC and was found to bind to the family member BRD7 with 10 fold lower affinity. The importance of chirality and configuration in this work is highlighted by the fact that the enantiomer of LP99, (2S, 3R)-LP99 showed no detectable binding to BRD9 by ITC.
This binding affinity was confirmed by HT-FRET where LP99 has an IC50 of 325nM
Selectivity
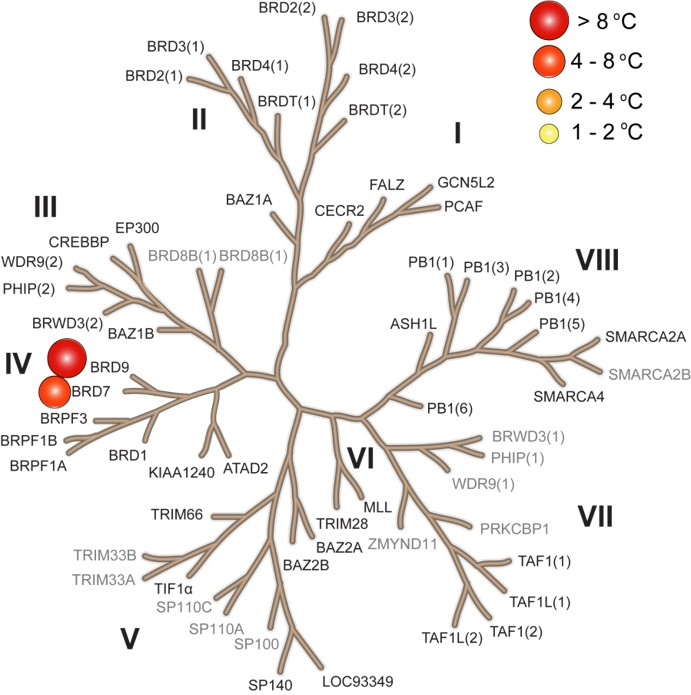
P. Clark et al, Angewandte Chemie Int. Ed. 2015
LP99 is a potent binder to BRD9 with a KD = 99 nM as shown by ITC and was found to bind to the family member BRD7 with 10 fold lower affinity. The importance of chirality and configuration in this work is highlighted by the fact that the enantiomer of LP99, (2S, 3R)-LP99 showed no detectable binding to BRD9 by ITC.
This binding affinity was confirmed by HT-FRET where LP99 has an IC50 of 325nM
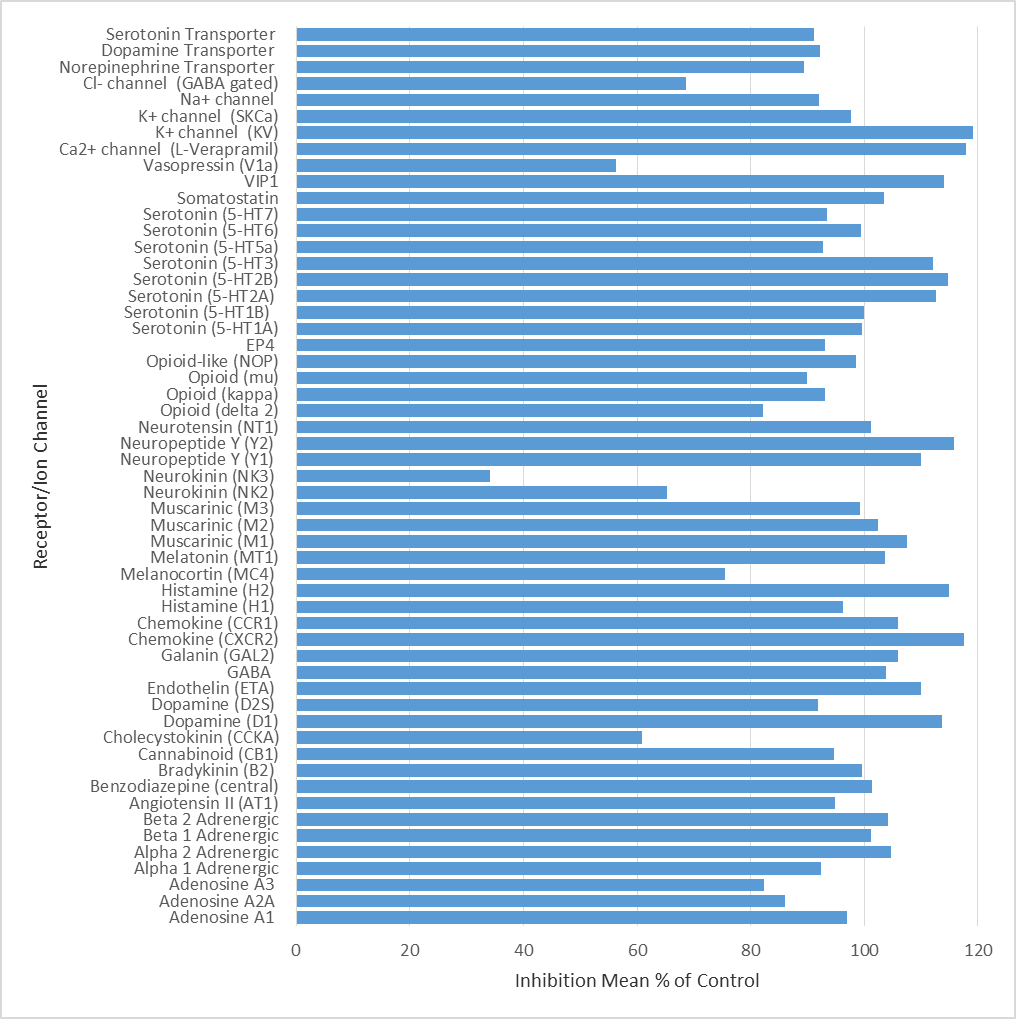
LP99 was found to be inactive vs 55 receptors and ion channels (CEREP panel) at 10 µM, 66% antagonist binding inhibition to NK3 was the most potent activity measured.
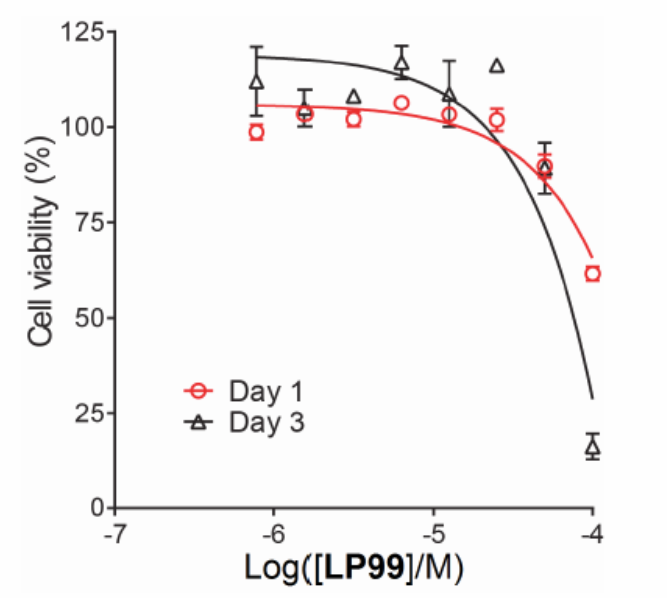
Cellular assays (FRAP, NanoBRET) demonstrate that the BRD7/9 inhibitor LP99 is able to disrupt the binding of BRD7 and BRD9 to chromatin in cells. Additionally, no cytotoxicity in U2OS cells was observed at 33 µM
 |
N-((2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl)-2-methylpropane-1-sulfonamide |
| Physical and chemical properties | |
|---|---|
| Molecular weight | 516.05 |
| Molecular formula | C26H30ClN3O4S |
| IUPAC name | N-((2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl)-2-methylpropane-1-sulfonamide |
| logP | 4.75 |
| PSA | 86.79 |
| No. of chiral centres | 2 |
| No. of rotatable bonds | 4 |
| No. of hydrogen bond acceptors | 9 |
| No. of hydrogen bond donors | 1 |
| Storage | Stable as solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | Soluble in DMSO at least up to ??? |
LP99 is a potent binder to BRD9 with a KD = 99 nM as shown by ITC. This binding is entirely driven by enthalpic interactions (ΔH=-11 kcal/mol), with a net loss in entropy upon binding (TΔS=-2.0 kcal/mol), which is consistent with a number of specific interactions with the protein. LP99 was found to bind to the family member BRD7 with 10 fold lower affinity. The importance of chirality and configuration in this work is highlighted by the fact that the enantiomer of LP99 showed no detectable binding to BRD9 by ITC.
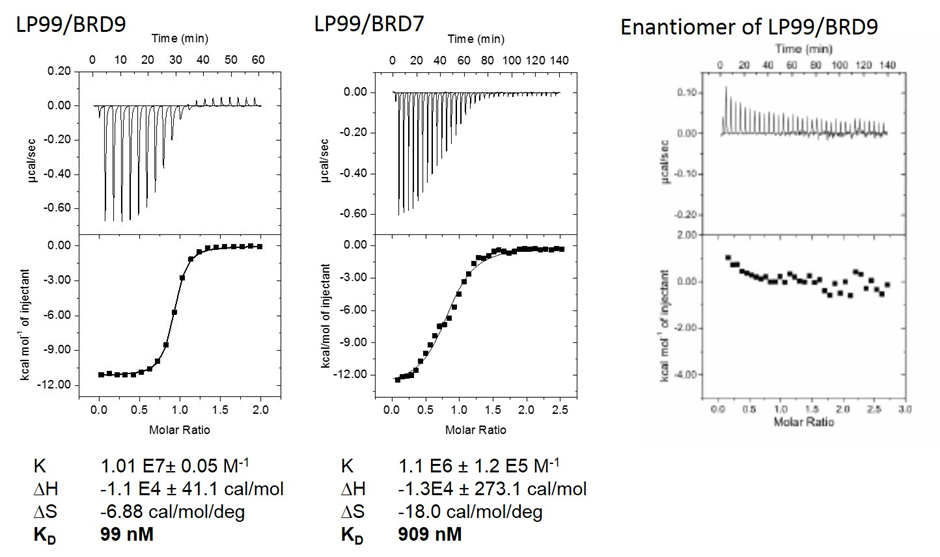
LP99 binds to BR9 with an IC50 of 325nM. The enantiomer (2S, 3R)-LP99 is inactive
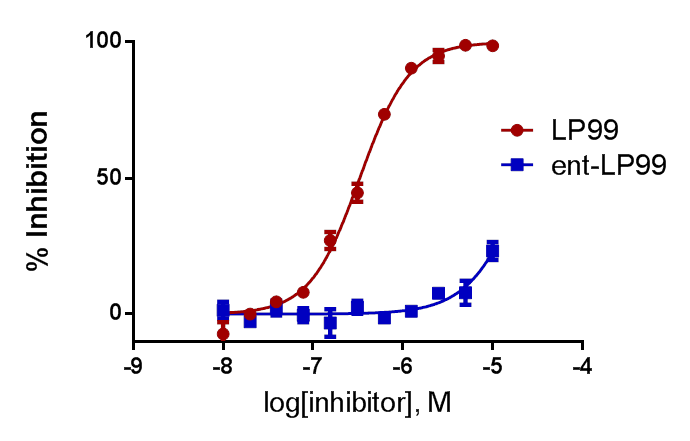
LP99 binds to BRD9 and BRD7 with high selectivity over other members of the Bromodomain family including members of the IV subfamily.
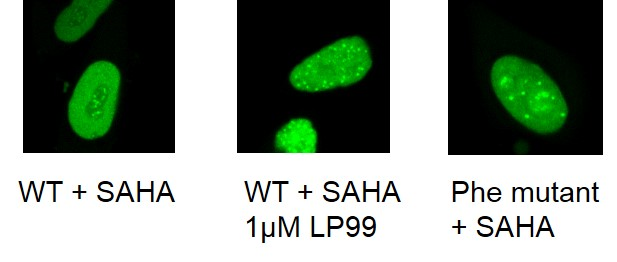
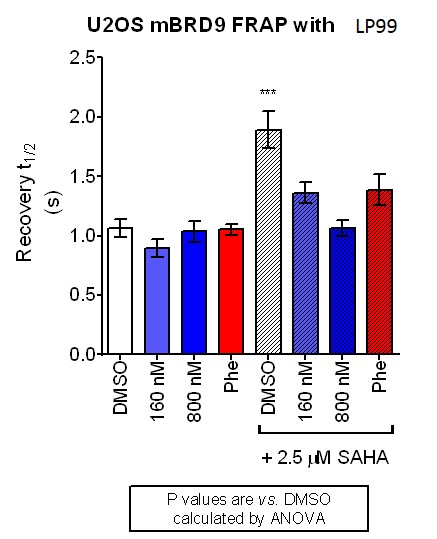
The cellular efficacy of LP99 on BRD9 was investigated using a fluorescence recovery after photo-bleaching (FRAP) assay [18]. LP99 was found to have a dose dependent effect on FRAP recovery time, with higher doses showing a t1/2 akin to that of cells expressing a N100F BRD9 mutant unable to bind histone proteins.
To measure this further, a bioluminescence resonance energy transfer (BRET) assay was performed. BRD7–and BRD9–NanoLuc luciferase fusion proteins and fluorescently labelled histone H3.3– and H4–HaloTag fusions were expressed in HEK293 cells.[19] The addition of LP99 decreased BRET for both BRD7 and BRD9 in both the H3.3 and H4 systems in a dose-dependent manner.
| Interaction | IC50 (uM) | Interaction | IC50 (uM) |
|---|---|---|---|
| BRD7-H3.3 | 3.7 ± 0.27 (4) | BRD9-H3.3 | 5.1 ± 0.50 (4) |
| BRD7-H4 | 3.3 ± 0.20 (4) | BRD9-H4 | 6.2 ± 0.59 (4) |
Cytotoxicity tests in U2OS cells for 24 and 72 hours showed the inhibitor to be non-toxic at concentrations of <33 uM.
Cytotoxicity tests in U2OS cells for 24 and 72 hours showed the inhibitor to be non-toxic at concentrations of <33 uM
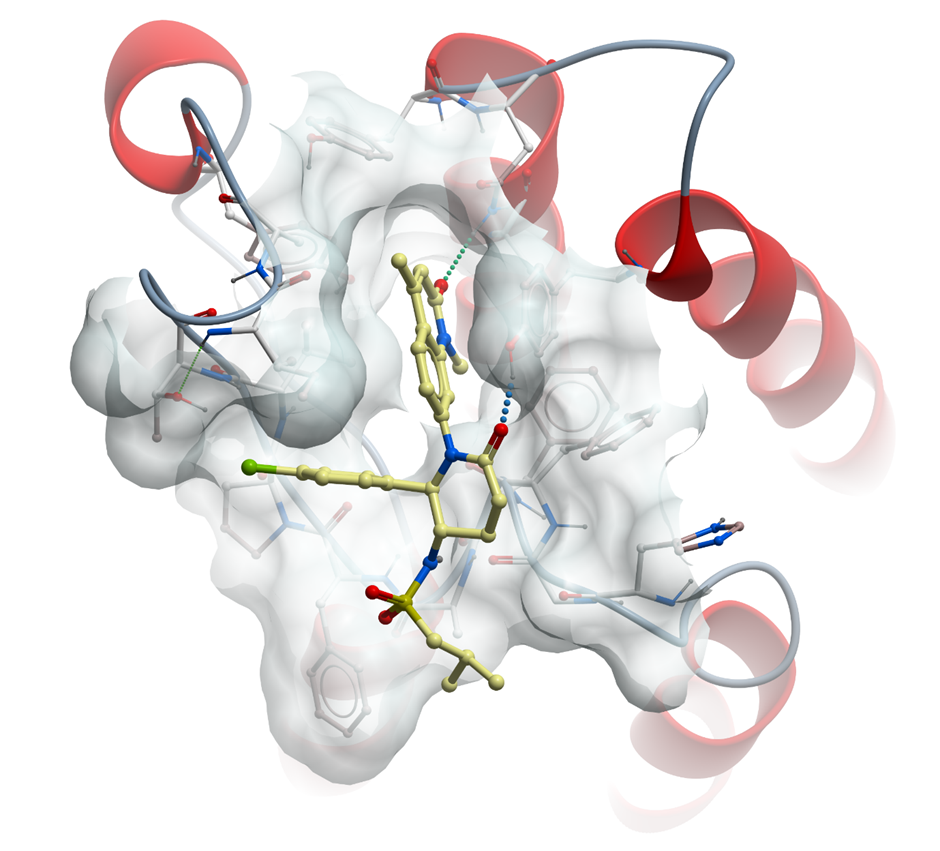
The co-crystal structure of LP99 with BRD9 has been solved with a resolution of 1.7 Å. The key features are
Isothermal Titration Calorimetry
Experiments were carried out on a VP-ITC titration microcalorimeter from MicroCalTM, LLC (Northampton, MA). All experiments were carried out at 15 °C while stirring at 295 rpm, in ITC buffer (50 mM HEPES pH 7.4 at 25 °C, 150 mM NaCl). The injection syringe (250 µl) was loaded with a solution of the protein sample (300 µM protein for the BETs, 950 µM protein for CREBBP and 600 µM for WDR9(2), in ITC buffer). All titrations were conducted using an initial injection of 2 µl followed by 34 identical injections of 8 µl with a duration of 16 sec (per injection) and a spacing of 250 sec between injections. The heat of dilution was determined by independent titrations (protein into buffer) and was subtracted from the experimental data. The collected data were implicated in the MicroCalTM Origin software supplied with the instrument to yield enthalpies of binding (ΔH) and binding constants (KB) as previously described by Wiseman and coworkers50. Thermodynamic parameters were calculated (ΔG = ΔH - TΔS = -RTlnKB, where ΔG, ΔH and ΔS are the changes in free energy, enthalpy and entropy of binding respectively). In all cases a single binding site model was employed.
TR-FRET BRD9 Binding Assay Using the EPIgeneousTM Binding Domain Kit B
This procedure is intended to measure the interaction between the BRD9 bromodomain and a peptide modified by acetylation of 4 lysine residues and can be used to assay compounds which bind to the bromodomain and inhibit this interaction. The assay uses a GST tagged BRD9 with anti-GST antibody conjugated Eu3+ cyrptate (donor) and a biotinylated peptide with Streptavidin conjugated XL-665 (acceptor). When the donor and acceptor labels are in close proximity, by binding of the peptide by the bromodomain, the excitation of the donor (337 nm) triggers a Fluorescence Resonance Energy Transfer (FRET) to the acceptor which then fluoresces at specific wavelength (665 nm)
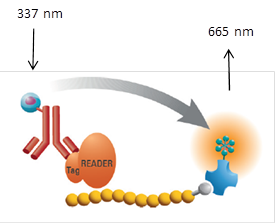
This assay was run using kit B from CisBio in accordance to manufacturer protocol.
strong>Differential Scanning Fluorimetry (DSF)
Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Stratagene). Proteins were buffered in 10 mM HEPES pH 7.5, 500 mM NaCl and assayed in a 96-well plate at a final concentration of 2 µM in 20 µl volume. Compounds were added at a final concentration of 10 µM. SYPRO Orange (Molecular Probes) was added as a fluorescence probe at a dilution of 1:1000. Excitation and emission filters for the SYPRO-Orange dye were set to 465 nm and 590 nm, respectively. The temperature was raised with a step of 3 °C per minute from 25 °C to 96 °C and fluorescence readings were taken at each interval. Data was analysed as previously reported [3]
CEREP assay
LP99 (10 µM) was screened against a panel of 55 ligand receptors, ion channels and transports using an established and widely utilized commercial assay platform (ExpresSProfile; CEREP, Paris, FRANCE).
Fluorescence Recovery After Photobleaching (FRAP) Assay
FRAP studies were performed using U20S cells expressing full-length BRD9 protein fused with an N-terminal eGFP as previously described [4]. Six hours after transfection 2.5uM SAHA (to increase global histone acetylation) was added and LP99 was added 1 hour before imaging, which was carried out 24 hours after transfection. Percent inhibition was calculated between the DMSO treated (0%) and N100F expressing mutant (100%)
NanoLuciferase Bioluminescent Resonance Energy Transfer (NanoBRET) Assay
HEK293 cells (8x105) were plated in each well of a 6-well plate and co transfected with one acceptor, Histone H3.3-HaloTag (NM_002107) or Histone H4 HaloTag (P62805); and one donor NanoLuc-BRD9 (Q9H8M2) BRD domain amino acids 120-240 or NanoLuc-BRD7 (NM_001173984) BRD domain amino acids 120-237. The cells were collected 20 hours after transfection, washed with PBS and exchanged into media containing phenol red-free DMEM and 4% FBS in the absence (control) or presence of 100nM NanoBRET 618 fluorescent ligand (Promega). Cell density was adjusted to 2x105 cells/ml and then re-plated into a 96-well white plate (Corning Costar #3917). LP99 was added at final concentrations between 0 and 33 uM and the plates incubated for 18hr at 37˚C in the presence of 5% CO2. NanoBRET furimazine substrate (Promega) was added at a final concentration of 10 uM. Readings were performed within 5 minutes using CLARIOstar (BMG) equipped with 450/80 nm bandpass and 610 nm longpass filters with a 0.5 s reading setting. A corrected BRET ratio was calculated (defined as ratio of emission at 610 nm/450 nm for samples treated with NanoBRET fluorescent ligand and emission at 610 nm/450 nm for samples not treated with NanoBRET fluorescent ligand). BRET ratios expressed as milli-BRET units (mBU) where 1 mBU corresponds to the corrected BRET ratio x 1000.
Cytotoxicity Assay
U20S cells were harvested from exponential phase cultures and plated in a 96-well opaque flat bottomed plates at a cell density of 3 x 103 cells/well (100uL). Compounds were dissolved in DMSO at 10 mM serial dilutions performed. 5 uL of compound solution was added to each well and incubated for 24 or 72 hours at 37˚C in a humidified atmosphere containing CO2 (5%). 10 uL of WST-1 (Roche) was added and the plates returned to the incubator. Plates were read on a plate reader at 450 nm after 2 h for cells treated with LP99 for 24 h or after 1 h for cells treated with LP99 for 72 h. Results plotted as % of DMSO control.