| Probe | Negative control | |
 |
|  |
NR162 |
| NR187 |
CASK (Ca2+/calmodulin-dependent Ser/Thr kinase) is a multidomain scaffolding protein, containing several protein-protein interaction domains. From the N-to the C-terminus CASK consists of a CAMK-like domain, two L27 domains, a PDZ domain, a SH3 domain and a guanylate kinase domain1. CASK is a member of the MAGUK (membrane-associated guanylate kinase) family that functions as a Mg2+-independent neurexin kinase with implicated roles in neuronal synapses and trafficking2. Furthermore, CASK has been categorized as a pseudokinase as it lacks the typical DFG motif and shows a GFG-motif instead. Pseudokinases carry a mutation at least in one of three highly conserved motifs within the catalytic kinase domain (VAIK-, DGF and Y/HRD motif) resulting in inactivation of catalytic activity. All in all 48 kinases of the kinome are pseudokinases3. However, CASK has been shown to maintain some catalytic activity in the absence of metal ions. High expression levels and mutations in CASK have been linked to diverse diseases, including colorectal cancer4, Parkinson's disease5 and X-linked mental retardation6, making CASK a potential drug target.
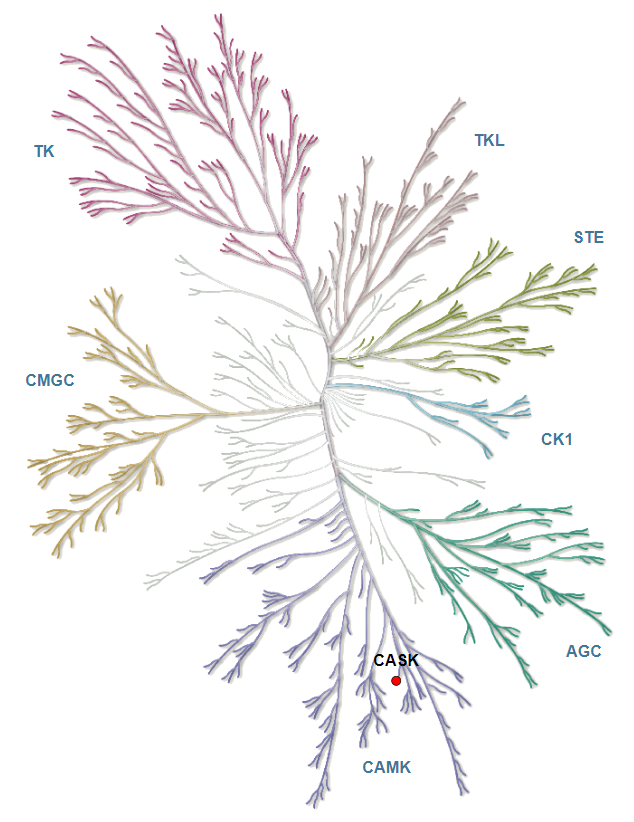
Phylogenetic kinase tree, CASK highlighted with red circle. Illustration is reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com).
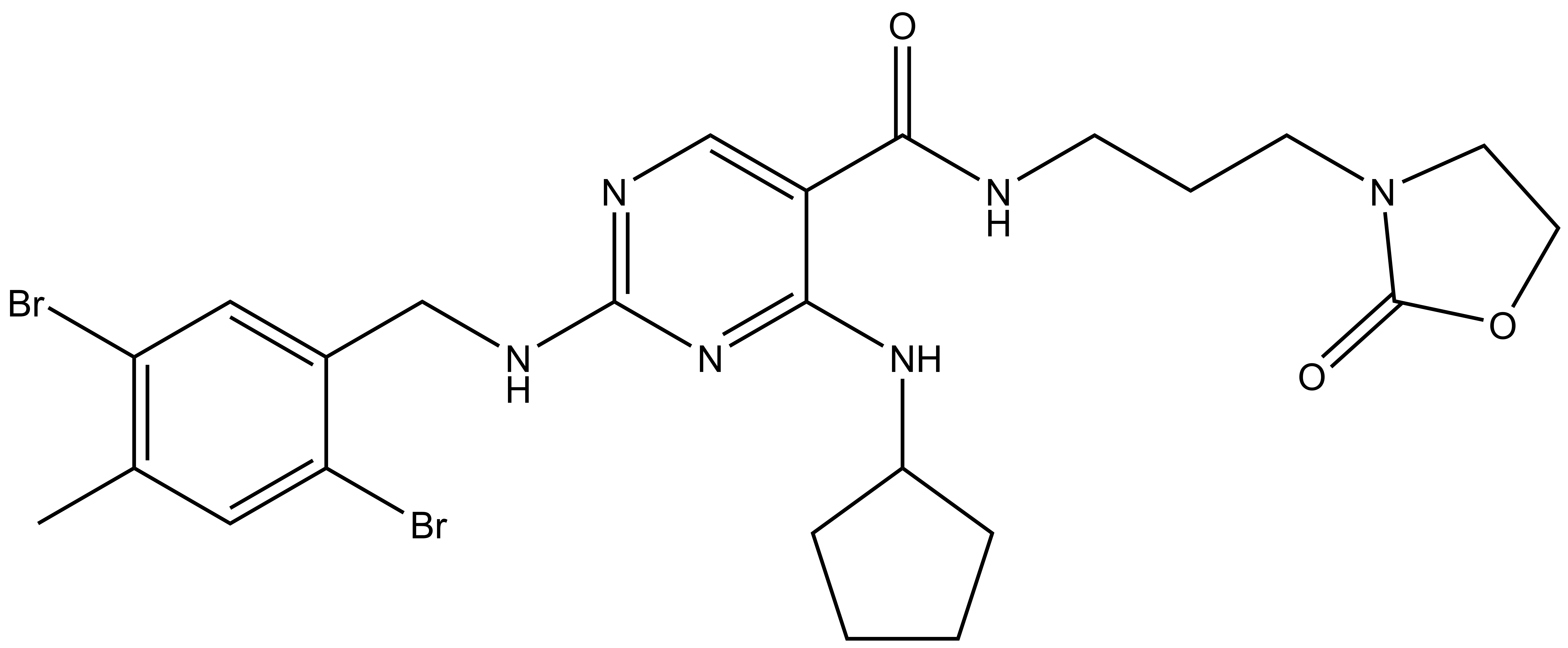
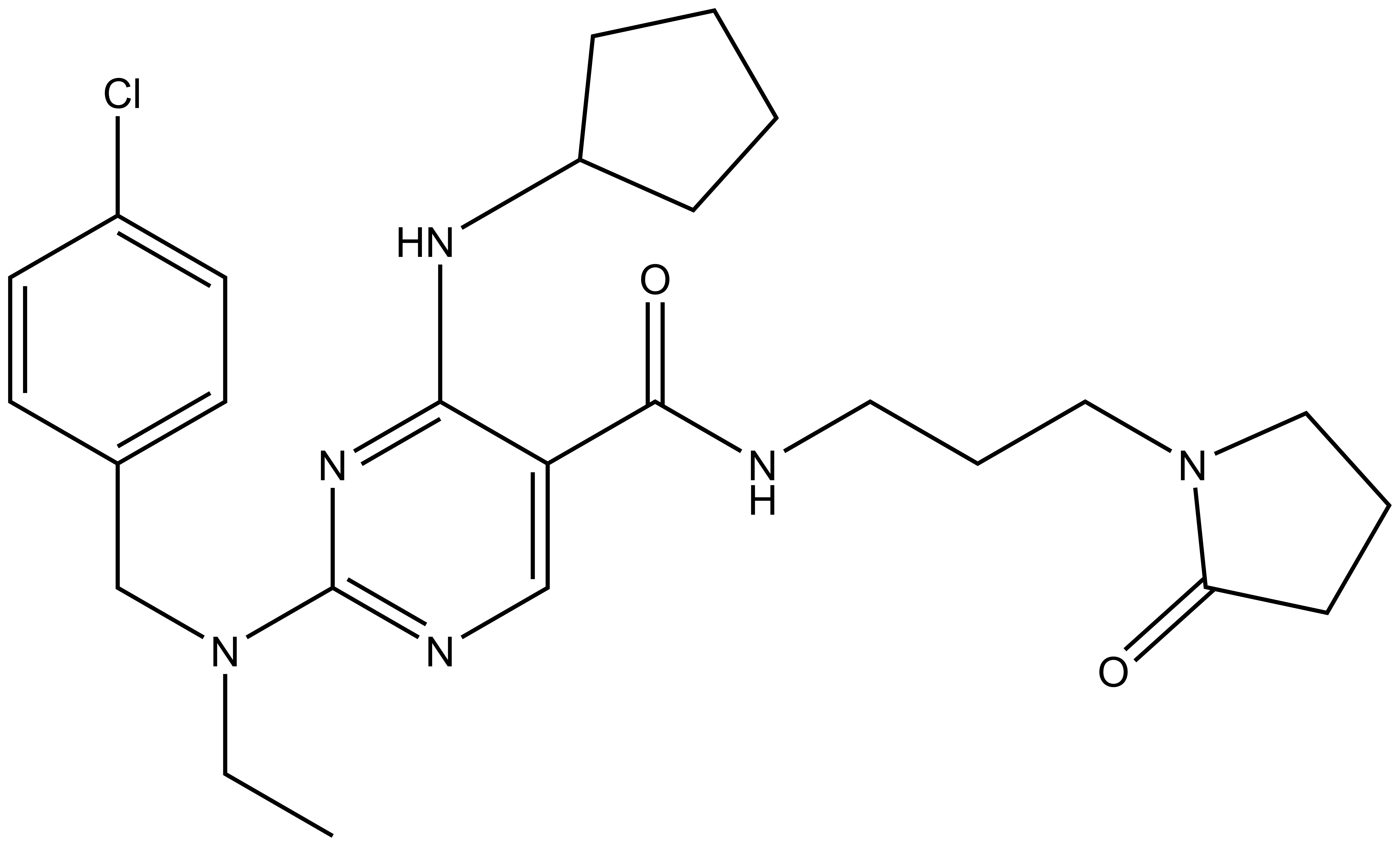
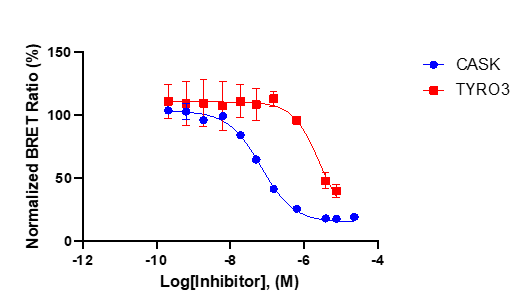
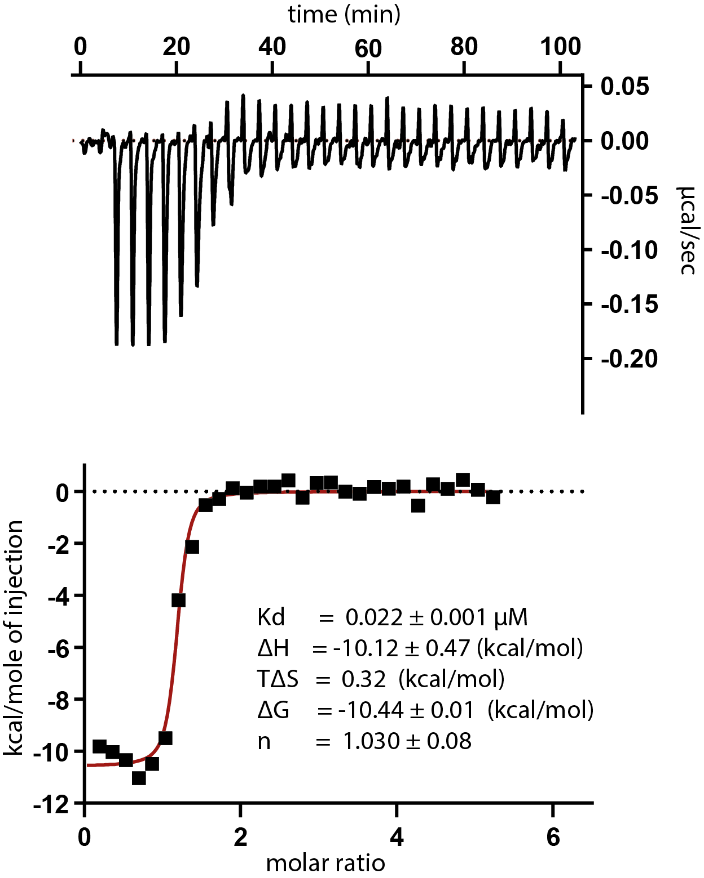
SGC has developed NR162, a potent and selective CASK inhibitor with a Kd of 22 nM in ITC and IC50 of 80 nM in NanoBRET assay. NR162 has been developed based on a series of TYRO3 inhibitors first published by Pfizer7. TYRO3 is a weak off target, but NR162 is 47-fold selective against TYRO3 (IC50 of 3.8 µM (NanoBRET). In addition, the chemical probe (NR162) is accompanied by a negative control (NR187), which is structurally similar to the probe molecule.
Potency Against Target Family
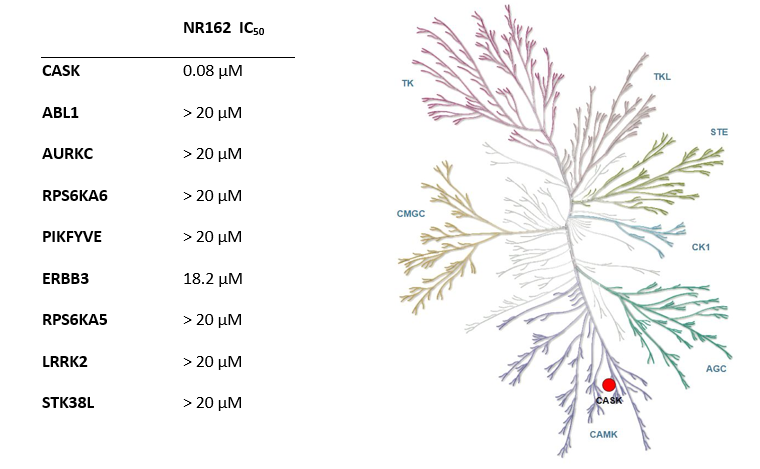
NR162 is a chemical probe for CASK with a Kd of 22 nM in ITC and an IC50 of 80 nM in NanoBRET assay. The closest off-target within the target family is ERBB3 with an IC50 of 18.2 µM (227 fold selective).
| Kinase | NR162 (µM) |
| CASK | 0.08 |
| ERBB3 | 18.2 |
Selectivity
NR162 has been shown to be selective in an in vitro kinase panel followed by cellular NanoBRET assays. Selectivity has been also confirmed by a DiscoverX KINOMEScan at 1000 nM. Off-targets were assessed as false positive in a follow-up NanoBRET. The selectivity outside target family revealed TYRO3 with an IC50 of 3.8 µM (47-fold) as closest off-target.
Dosage
To minimize the chance of off-target effects, we recommend a concentration of no higher than 10 µM for cell-based assays.
Cellular Activity
In NanoBRET assay using HEK923T cells NR162 shows an IC50 of 80 nM against on target.
| NR162 |
 |
Click here to download the SDF file. |
| NR162 | |
| Physical and chemical properties | |
| Molecular weight | 608.07 |
| Molecular formula | C24H30Br2N6O3 |
| IUPAC name | 4-(cyclopentylamino)-2-((2,5-dibromo-4-methylbenzyl)amino)-N-(3-(2-oxooxazolidin-3-yl)propyl)pyrimidine-5-carboxamide |
| logP | 4.2 |
| TPSA | 107.42 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 10 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 3 |
| Storage | stable as solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | soluble in DMSO |
| NR187 |
 |
Click here to download the SDF file. |
| NR187 | |
| Physical and chemical properties | |
| Molecular weight | 499.06 |
| Molecular formula | C26H35ClN6O2 |
| IUPAC name | 2-((4-chlorobenzyl)(ethyl)amino)-4-(cyclopentylamino)-N-(3-(2-oxopyrrolidin-1-yl)propyl)pyrimidine-5-carboxamide |
| clogP | 3.53 |
| TPSA | 89.4 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 11 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 2 |
| Storage | stable as solid in the dark at -20°C. NB making aliquots rather than freeze-thawing is recommended |
| Dissolution | soluble in DMSO |
SMILES:
NR162: O=C(C1=CN=C(N=C1NC2CCCC2)NCC3=CC(Br)=C(C=C3Br)C)NCCCN4CCOC4=O
NR187: O=C(C1=CN=C(N=C1NC2CCCC2)N(CC3=CC=C(C=C3)Cl)CC)NCCCN4CCCC4=O
InChI:
NR162: InChI=1S/C24H30Br2N6O3/c1-15-11-20(26)16(12-19(15)25)13-28-23-29-14-18(21(31-23)30-17-5-2-3-6-17)22(33)27-7-4-8-32-9-10-35-24(32)34/h11-12,14,17H,2-10,13H2,1H3,(H,27,33)(H2,28,29,30,31)
NR187: InChI=1S/C26H35ClN6O2/c1-2-32(18-19-10-12-20(27)13-11-19)26-29-17-22(24(31-26)30-21-7-3-4-8-21)25(35)28-14-6-16-33-15-5-9-23(33)34/h10-13,17,21H,2-9,14-16,18H2,1H3,(H,28,35)(H,29,30,31)
InChIKey:
NR162: BPLPIBNWDLPUKP-UHFFFAOYSA-N
NR187: AYRUNBYNKCLGFI-UHFFFAOYSA-N
A DiscoverX KINOMEScan of NR162 at 1000 nM revealed very few off-targets.
| Target | NR162 (% Control) |
| CASK | 0 |
| AURKC | 0 |
| ABL1 | 0 |
| RPSKA6 | 15 |
| PIKFYVE | 30 |
| ERBB3 | 35 |
| RPS6KA5 | 35 |
| LRRK2 | 41 |
| STK38L | 42 |
Hits from the KINOMEScan were assessed in a cellular NanoBRET assay in HEK293T cells and IC50 of 18.2 µM for ERBB3 (227 fold selective) was determined. All other off-targets were not confirmed in the follow-up NanoBRET assay.
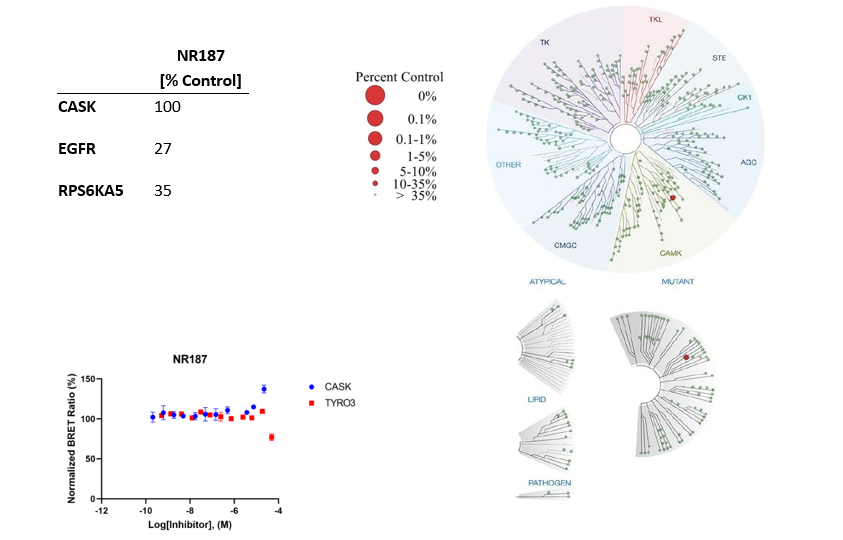
The negative control NR187 with its blocked hinge-binding amine showed no activity in NanoBRET and KINOMEScan was also clean.
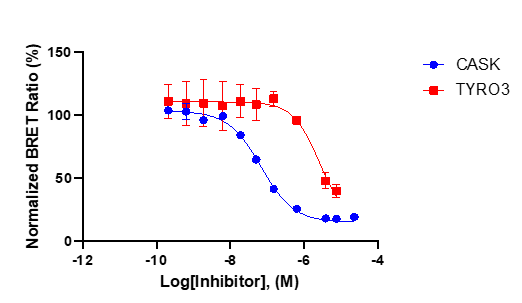
In NanoBRET assay perfomed in HEK923T cells NR162 displayed the following IC50 value against CASK 80 nM and 3.8 µM against TYRO3.
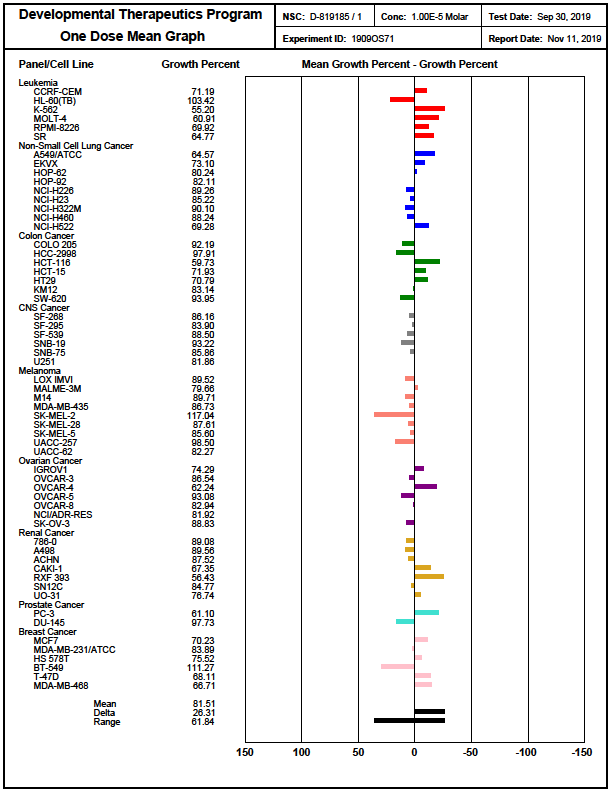
In the NCI-60 screen, which is a human tumor cell line screen, NR162 showed no significant cell toxicity as well as growth inhibition. It was tested in a single high dose of 10 µM in the full NCI-60 panel. https://dtp.cancer.gov/discovery_development/nci-60/methodology.htm
Crystallography revealed that due to the missing aspartate in CASK the inhibitor shows an improved selectivity pattern and is no longer active on MERTK, AXL and ABL1 and affinity is reduced to TYRO3 (off-targets of the lead structure PFE-PKIS12).
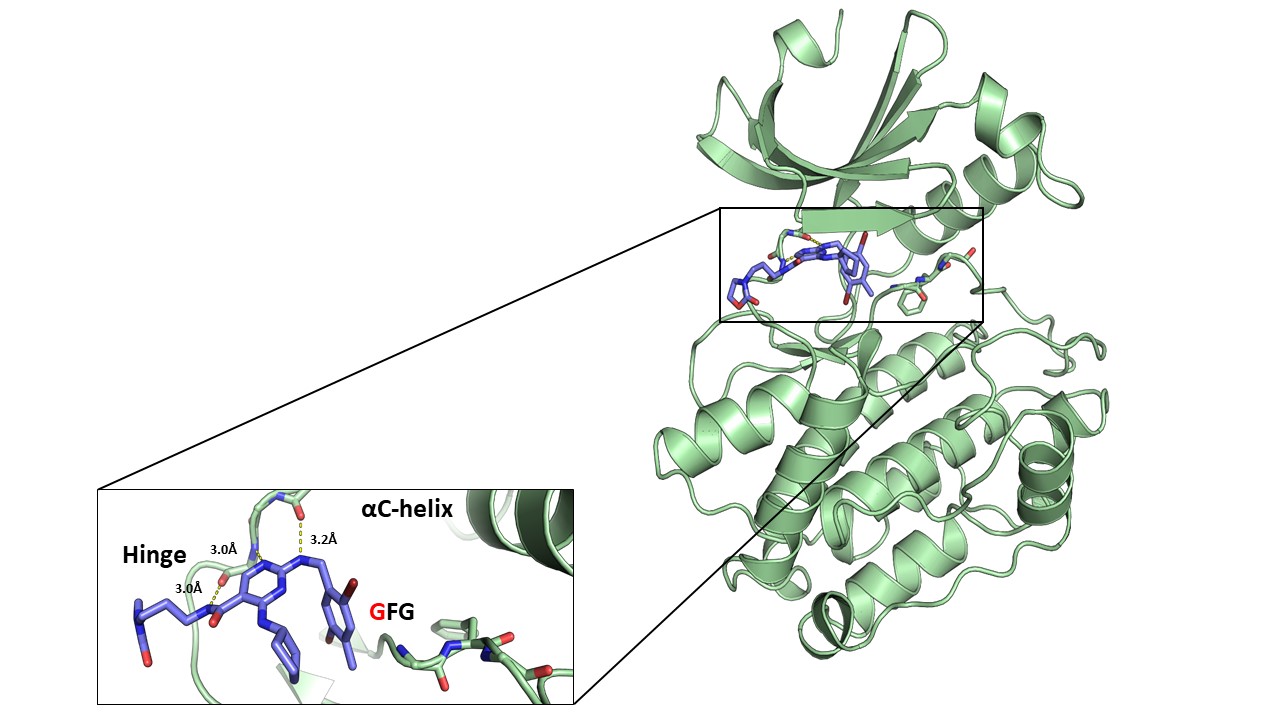
Figure 1: Overview of NR162 in complex with CASK (right) and details of the interaction of NR162 with the CASK hinge region (left).
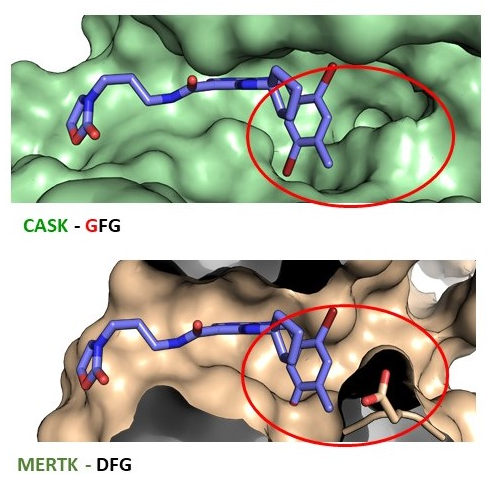
Figure 2: Co-crystal structure of NR26_162 in complex with CASK (GFG-pocket) and alignment with MERTK (DFG-pocket).
DSF assay
Recombinant proteins were assessed as described in (Fedorov O, Niesen FH, Knapp S. Kinase inhibitor selectivity profiling using differential scanning fluorimetry. Methods Mol Biol. 2012;795:109-18.)
ITC
The ITC measurement was performed on a NanoITC (TA Instruments) at 25°C in buffer containing 30 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM TCEP and 5% Glycerol. CASK at 141 µM was injected into the cell, containing compounds at 2-6 µM. The integrated heat of titration was calculated and fitted to a single, independent binding model using the software provided by the manufacture. The thermodynamic parameters (ΔH and TΔS), equilibrium association and dissociation constants (Ka and KD), and stoichiometry (n) were calculated.
NanoBRET (Promega)
N-terminal NanoLuc and C-terminal NanoLuc/ kinase fusions, encoded in pFC32K expression vectors (Promega), were used. For cellular BRET target engagement experiments, HEK-293T were transfected with NLuc/target fusion constructs using FuGENE HD (Promega) according to the manufacturer’s protocol. Briefly, Nluc/target fusion constructs were diluted into Transfection Carrier DNA (Promega) at a mass ratio of 1:10 (mass/mass), diluted with OptiMEM media to a twentieth part of the volume of the HEK cells. FuGENE HD was added at a ratio of 1:3 (μg DNA: μL FuGENE HD). One part (vol) of FuGENE HD complexes was combined with 20 parts (vol) of HEK-293 cells suspended at a density of 2 x 105 cells/ml and afterwards the mixture was incubated in a humidified, 37°C/5% CO2 incubator for 24 h.
After this, the cells were washed and resuspendend in OptiMEM medium. For Target engagement assays 4 x 103 cells/well were plated out in a 384-well plate (Greiner). For all experiments the recommended energy transfer probes (Promega) were used if possible, at a final concentration of the Kd of the tracer on the target. In some cases, higher tracer concentrations were used for better signal-to-noise ratios. Compounds and the energy transfer probe were added to the cells and incubated for 2h in humidified, 37°C/5% CO2 incubator. The chemical inhibitors were prepared as concentrated stock solutions in DMSO (Sigma-Aldrich) and diluted with OptiMEM for this experiment. Straight before the measurement NanoGlo Substrate and Extracellular NanoLuc Inhibitor (Promega) were mixed carefully with the supernatant.
Luminescence was measured on a BMP PheraStar with 450 nm (donor) and 600 nm filters (acceptor) using 0.5 s integration time. Milli-BRET units (mBU) are calculated by multiplying the raw BRET values by 1000. Tracer and DMSO controls were used to calculate a normalized signal.
mBU=1000*I[600 nm]/I[450 nm]
Inhibitory constants were calculated by using the sigmoidal dose-response (four parameters) equation in GraphPad Prism.
Y=Bottom+((Top-Bottom) )/(1+〖10〗^(X-LogIC50*HillSlope) )