| Probe | Negative control | |
 |
|  |
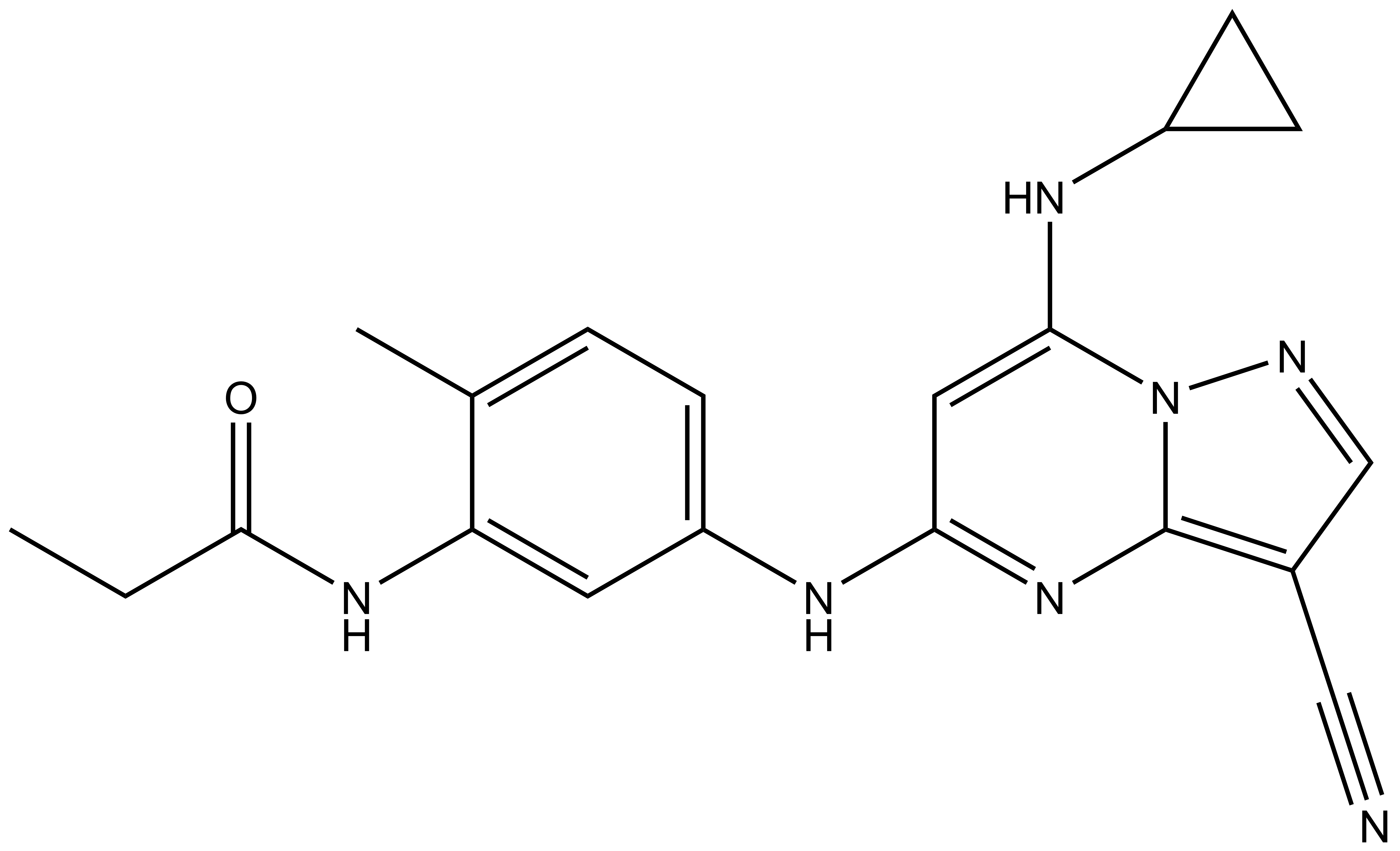
SGC-CK2-1 |
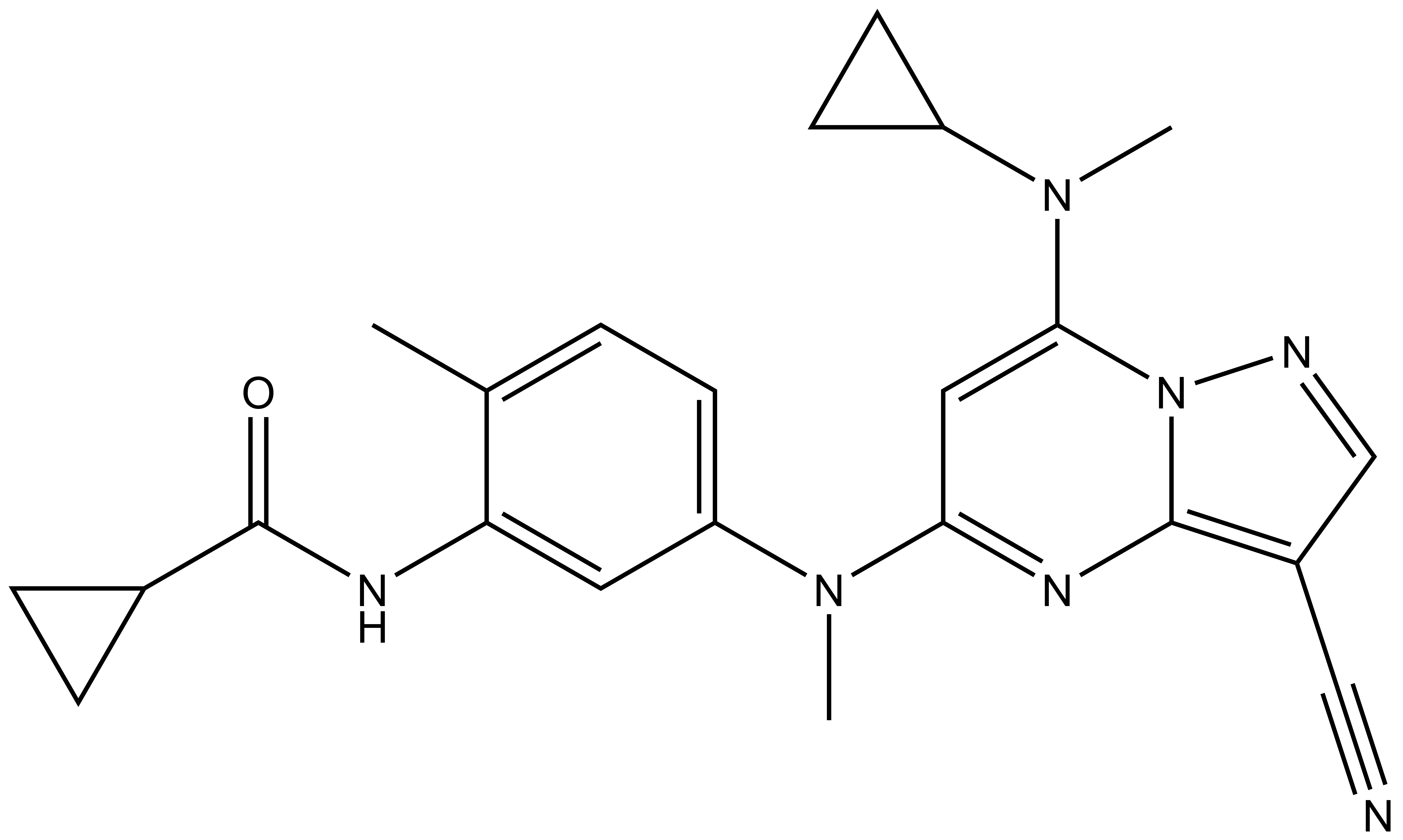
| SGC-CK2-1N |
Based upon a pyrazolopyrimidine scaffold with potent CK2 inhibition exemplified by AstraZeneca, the SGC has developed a high-quality chemical probe and its negative control for protein kinase CK2.1 This compound shows equal potency in binding to both catalytic subunits of CK2: CK2α (CSNK2A1) and CK2α' (CSNK2A2). CK2 is a serine/threonine kinase that is part of the larger CMGC family, which is named after the initials of its subfamily members including cyclin-dependent kinases (CDK), mitogen-activated protein kinase (MAPK), glycogen synthase kinase (GSK), and CDC-like kinase (CLK). A plethora of biological functions have been ascribed to CK2 ranging from cell survival and proliferation to inflammation. CK2 phosphorylates more than 300 proteins and, via genetic and biochemical studies in a variety of experimental models, has been found to be both constitutively active and ubiquitously expressed.2-5 While CK2 inhibition has been widely studied in the context of cancer, pervasive use of non-selective CK2 inhibitors has confounded interpretation of results in many oncological studies.
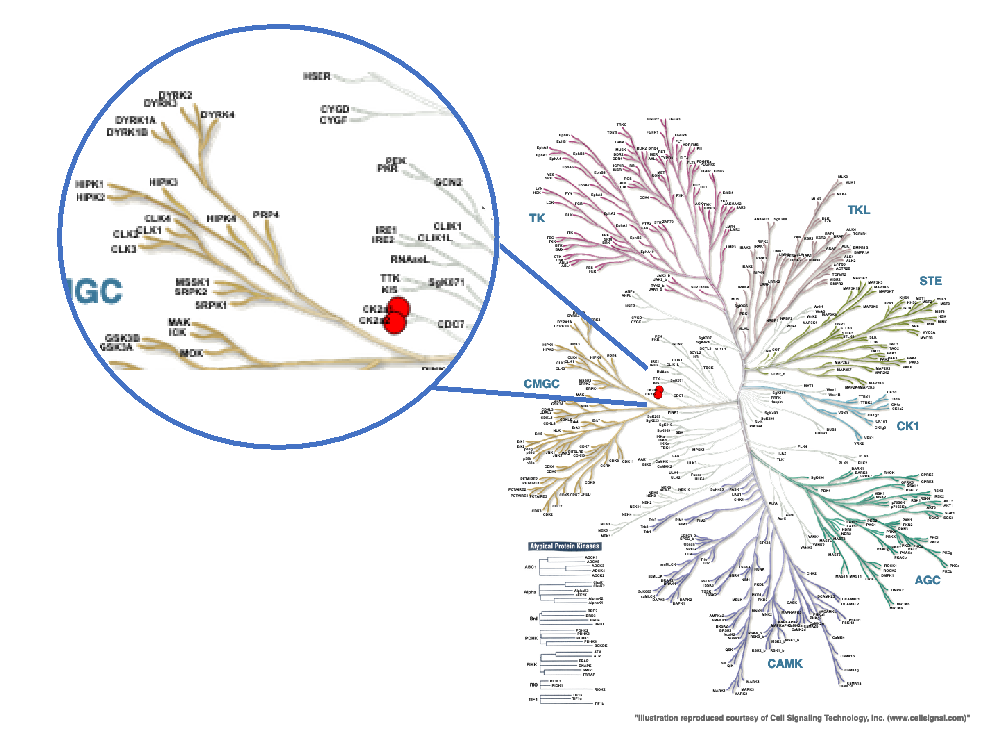
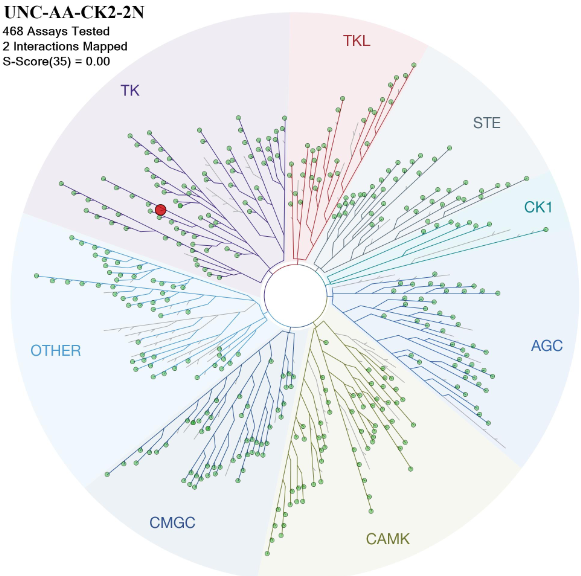
Figure 1: Phylogenetic kinase tree with CK2α and CK2α' highlighted with red circles. Illustration is reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com)
Key profiling data:
| Probe |
 |
SGC-CK2-1 |
SMILES:
CC1=CC=C(C=C1NC(CC)=O)NC2=NC3=C(C=NN3C(NC4CC4)=C2)C#N
InChI:
InChI=1S/C20H21N7O/c1-3-19(28)25-16-8-15(5-4-12(16)2)23-17-9-18(24-14-6-7-14)27-20(26-17)13(10-21)11-22-27/h4-5,8-9,11,14,24H,3,6-7H2,1-2H3,(H,23,26)(H,25,28)
InChIKey: YKDZIFFKQUNVHH-UHFFFAOYSA-N
| Physical and chemical properties | |
| Molecular weight | 375.44 |
| Molecular formula | C20 H21 N7 O |
| IUPAC name | N-(5-((3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl)amino)-2-methylphenyl)propionamide |
| clogP | 1.94 |
| PSA | 104.91 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 7 |
| No. of hydrogen bond acceptors | 4 |
| No. of hydrogen bond donors | 3 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10 mM) are stable at -20oC |
| Dissolution | Soluble up to 10mM in DMSO |
| Negative control |
 |
SGC-CK2-1N |
SMILES:
CC1=CC=C(C=C1NC(C2CC2)=O)N(C3=NC4=C(C=NN4C(N(C5CC5)C)=C3)C#N)C
InChI:
InChI=1S/C23H25N7O/c1-14-4-7-18(10-19(14)26-23(31)15-5-6-15)28(2)20-11-21(29(3)17-8-9-17)30-22(27-20)16(12-24)13-25-30/h4,7,10-11,13,15,17H,5-6,8-9H2,1-3H3,(H,26,31)
InChIKey: LGMBLPOARZRSKR-UHFFFAOYSA-N
| Physical and chemical properties | |
| Molecular weight | 415.2 |
| Molecular formula | C23 H25 N7 O |
| IUPAC name | 4-((3-(cyclopropyl-formylamino)-4-methyl-phenyl)-methyl-amino)-2-(cyclopropyl-methyl-amino)-1,5,9-triaza-bicyclo[4.3.0]nona-2,4,6,8-tetraene-7-carbonitrile |
| clogP | 3.67 |
| PSA | 63.46 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 8 |
| No. of hydrogen bond acceptors | 5 |
| No. of hydrogen bond donors | 1 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10 mM) are stable at -20oC |
| Dissolution | Soluble up to 10mM in DMSO |
SGC-CK2-1 was profiled in the KINOMEscan assay against 403 wild-type kinases at 1 μM. Only 11 kinases showed PoC <35 giving an S(35) at 1 μM = 0.027. Potential off-targets were tested in the nanoBRET target engagement assay (DYRK2) and via Eurofins radiometric and LANCE kinase assays. Data corresponding with off-target kinase activity is shown in the table below.
| Kinase | % Control at 1µM | Enzymatic IC50 (nM) | NanoBRET IC50 (nM) |
| CK2α'/CSNK2A2 | 0 | 2.3 | 16 |
| DRAK1 | 0 | >10000 | NT |
| CK2α/CSNK2A1 | 0.5 | 4.2 | 36 |
| DYRK2 | 14 | 440 | 3700 |
| PLK4 | 23 | >10000 | NT |
| HIPK2 | 26 | 3400 | NT |
| MEK5 | 28 | 0% AT 1µM | NT |
| HIPK1 | 32 | 3700 | NT |
| HIPK3 | 34 | 8100 | NT |
| RIOK2 | 32 | NT | NT |
| SGK3 | 34 | >10000 | NT |
Figure 1: SGC-CK2-1 was profiled in the KINOMEscan assay against 403 wild-type kinases at 1 μM and off-target kinases inhibited PoC <35 were tested in an orthogonal assay.
SGC-CK2-1N was also tested in the DiscoverX panel and 1 kinase had a PoC <35. The negative control was sent to Eurofins for testing in enzyme assays for CSNK2A1 and CSNK2A2. The results are in the table below.
| Kinase | % Control at 1µM | Enzymatic IC50 (nM) | NanoBRET IC50 (nM) |
| RIOK1 | 23 | NT | NT |
| RET | 35 | NT | NT |
| FLT3 | 40 | NT | NT |
| BRAF | 55 | NT | NT |
| IKK-alpha | 58 | NT | NT |
| MKNK1 | 58 | NT | NT |
| STK33 | 61 | NT | NT |
| TNK1 | 62 | NT | NT |
| EPHA2 | 63 | NT | NT |
| CK2α'/CSNK2A2 | 65 | >10000 | >10000 |
| CK2α/CSNK2A1 | 100 | >10000 | NT |
Figure 2: SGC-CK2-1N was profiled in the KINOMEscan assay against 403 wild-type kinases at 1 μM and follow-up CK2α and CK2α' enzymatic assays were done to confirm no activity.
A NanoBRET assay was utilized to assess the binding affinity of SGC-CK2-1 to CK2α and CK2α'. The negative control shows no binding affinity for CK2α'.
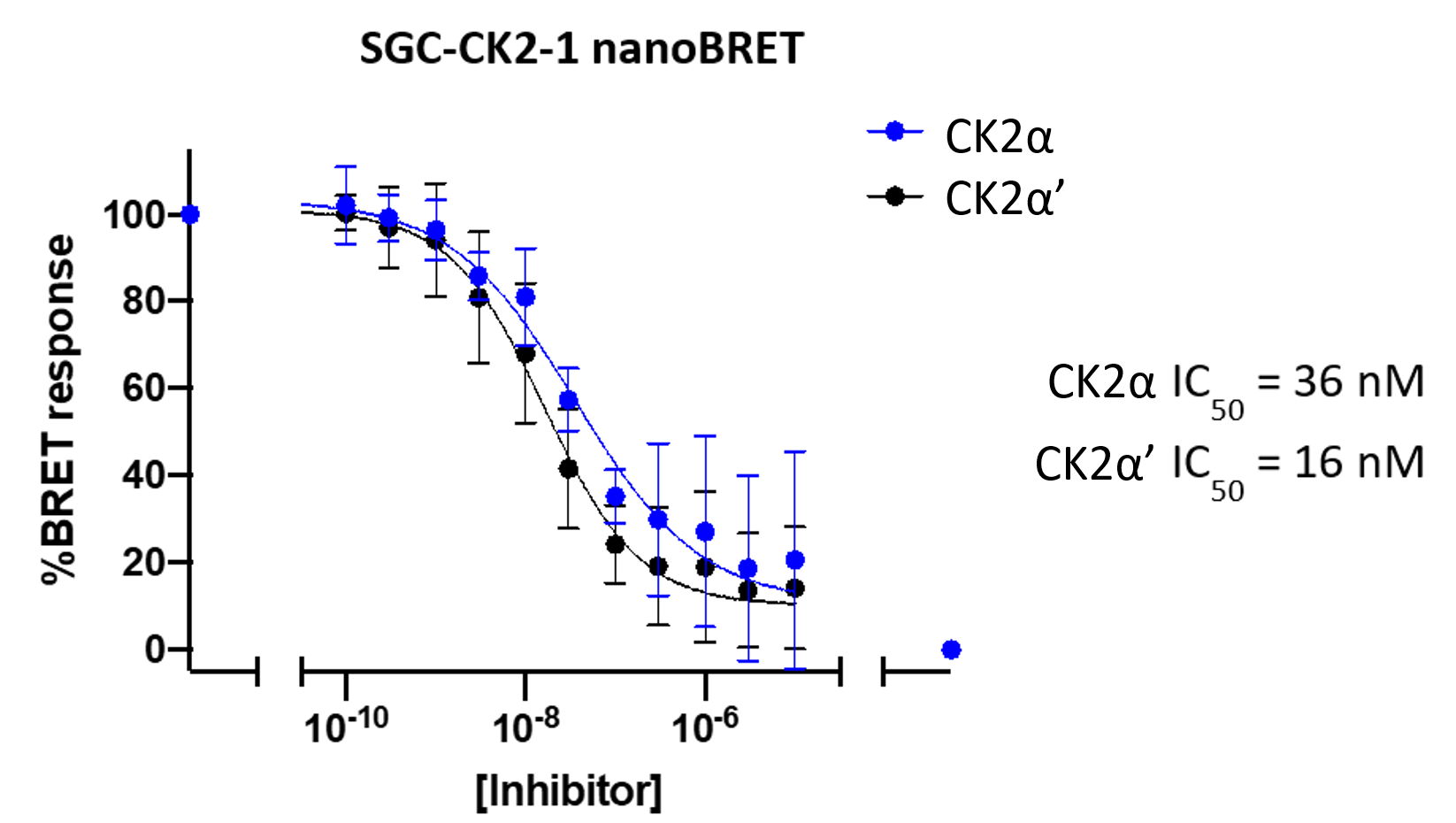
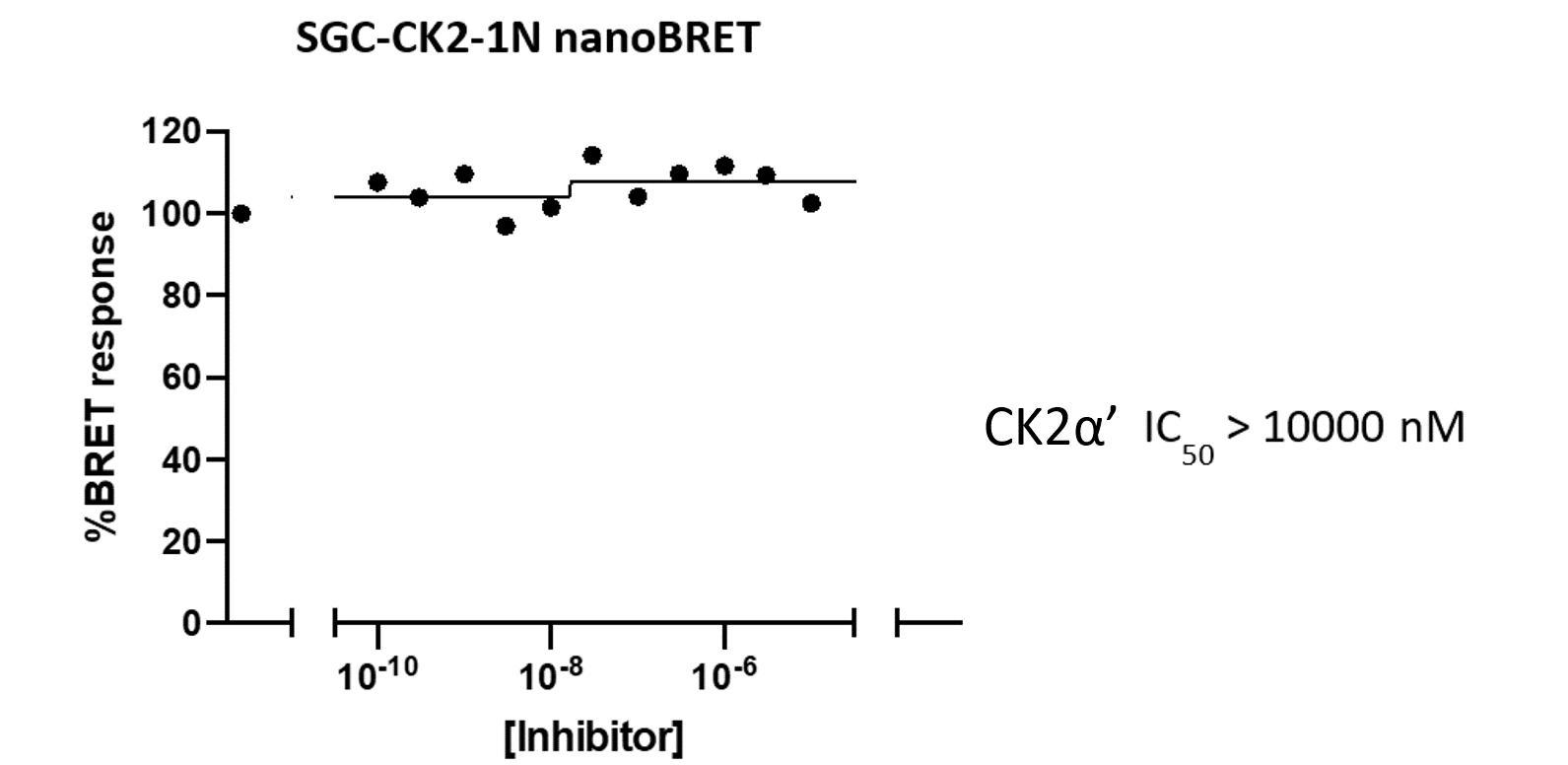
Figure 1: SGC-CK2-1 and SGC-CK2-1N were profiled in the CK2 NanoBRET assays.
Related Links: https://www.tocris.com/products/sgc-ck2-1_7450#product-literature