| Probe | Negative control | |
 |
|  |
SGC-STK17B-1 |
| SGC-STK17B-1N |
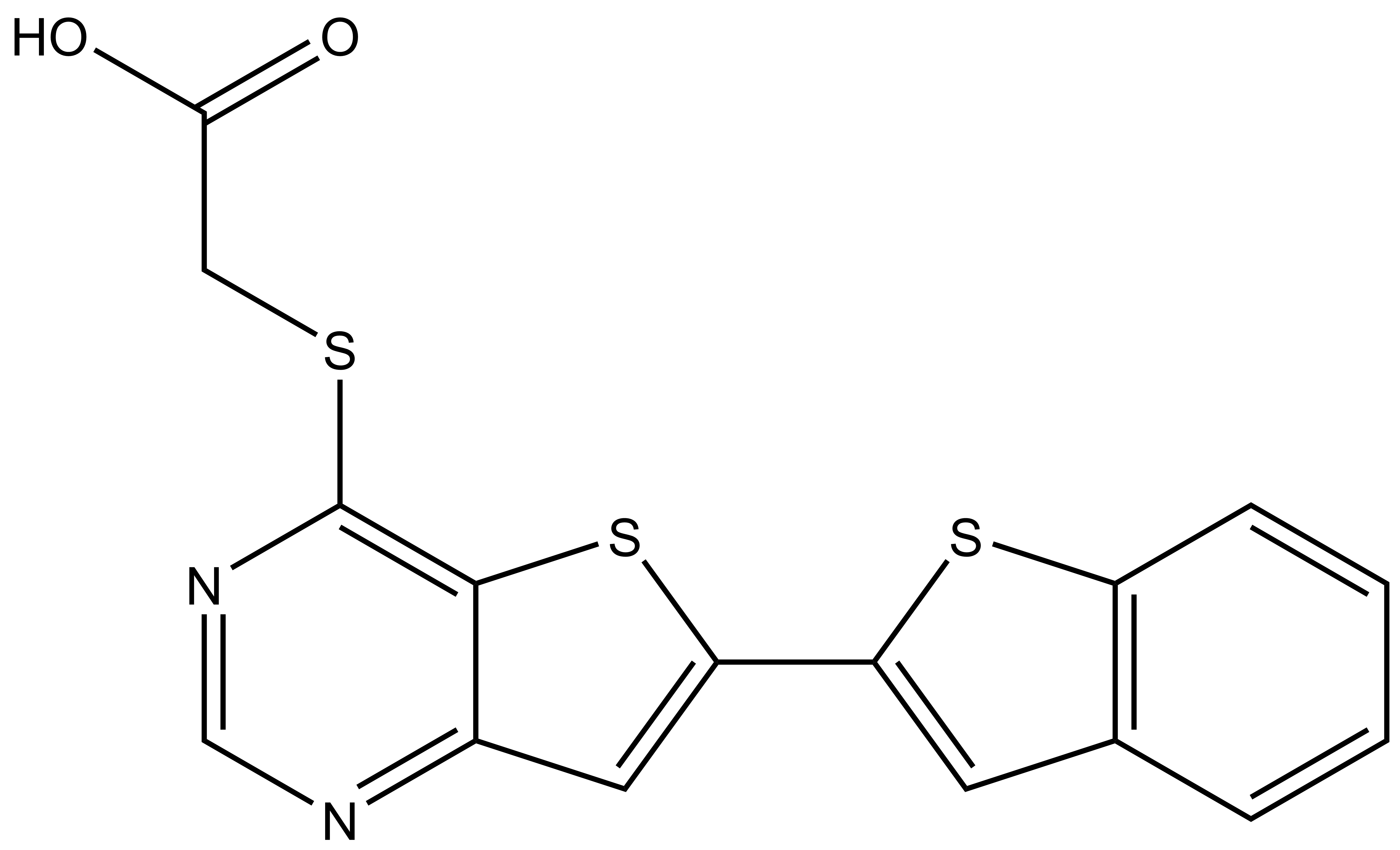
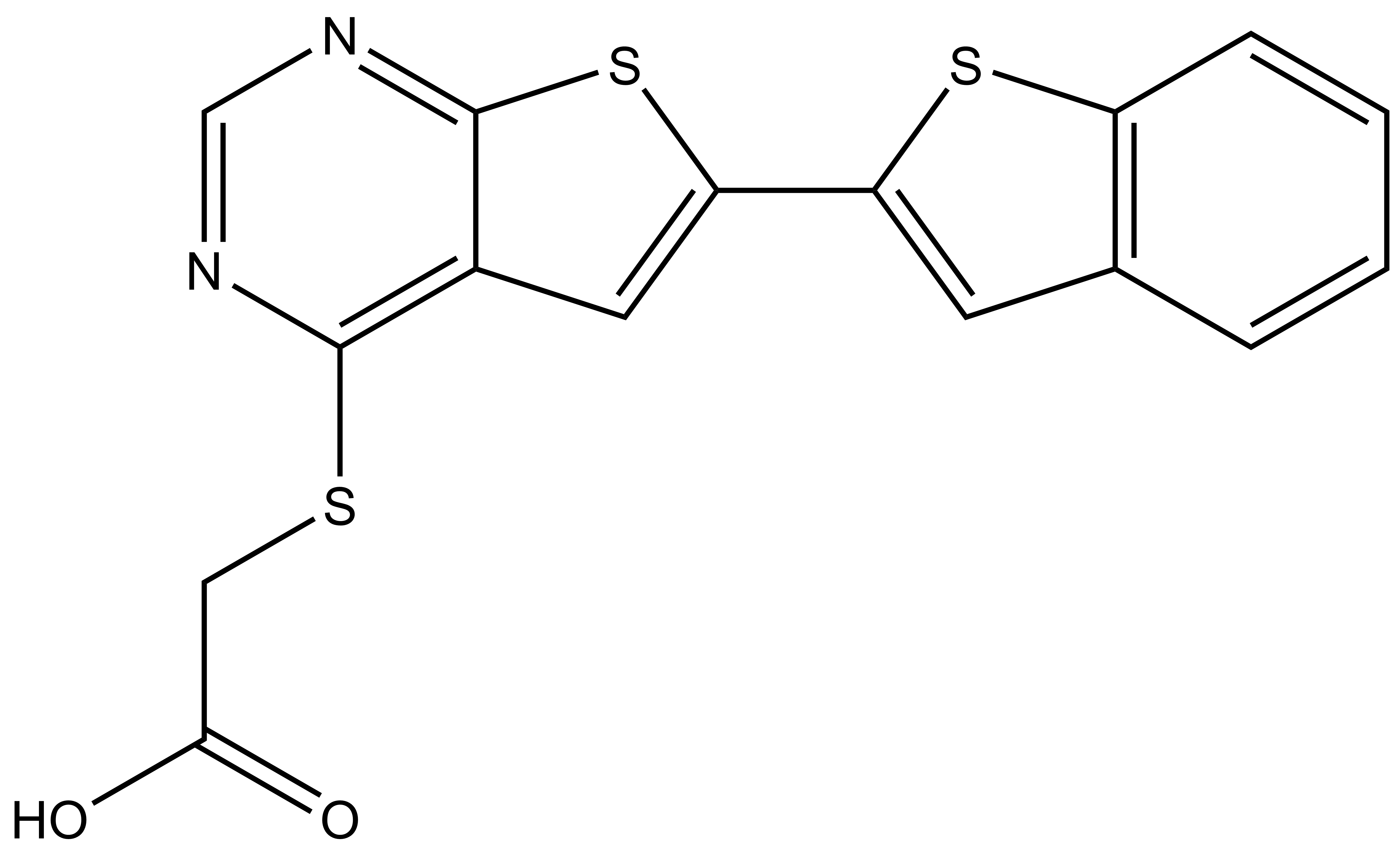
Chemical structures of SGC-STK17B-1 (thieno[3,2-d]pyrimidine) and the negative control SGC-STK17B-1N (thieno[2,3-d]pyrimidine).
In a collaboration with Pfizer, the SGC has developed a high quality chemical probe and its negative control for protein kinase STK17B/DRAK2, Figure 1, which is a serine/threonine kinase part of the larger Death-associated protein kinase (DAPK) family that also includes DAPK1, DAPK2, DAPK3, and DRAK1. STK17B kinase is predominantly expressed in T-cells and B-cells. Due to this expression it has been identified as a possible therapeutic target for Type 1 diabetes, Multiple sclerosis and graft rejection. STK17B knock out mice are viable and show no abnormalities, but are remarkably resistant to autoimmune challenge. When crossed onto the NOD1 strain, a model of Type 1 diabetes1 or challenged with EAE, a model of Multiple Sclerosis, the STK17B knock out mice are protected from disease progression.2 STK17B is also overexpressed in hepatocellular3 and breast cancer.4 Silencing of STK17B in cells suggests that it may have utility in treatment of these cancers. The starting point in the development of this chemical probe was a series of thienopyrimidines donated by Pfizer, prepared in an effort targeting TPL2 kinase.5 From here, the SGC developed a library of small molecules inhibitors around the thieno[3,2-d]pyrimidine core optimizing for potency, selectivity, and cell activity and from this set, we identified SGC-STK17B-1 as a small molecule chemical probe.
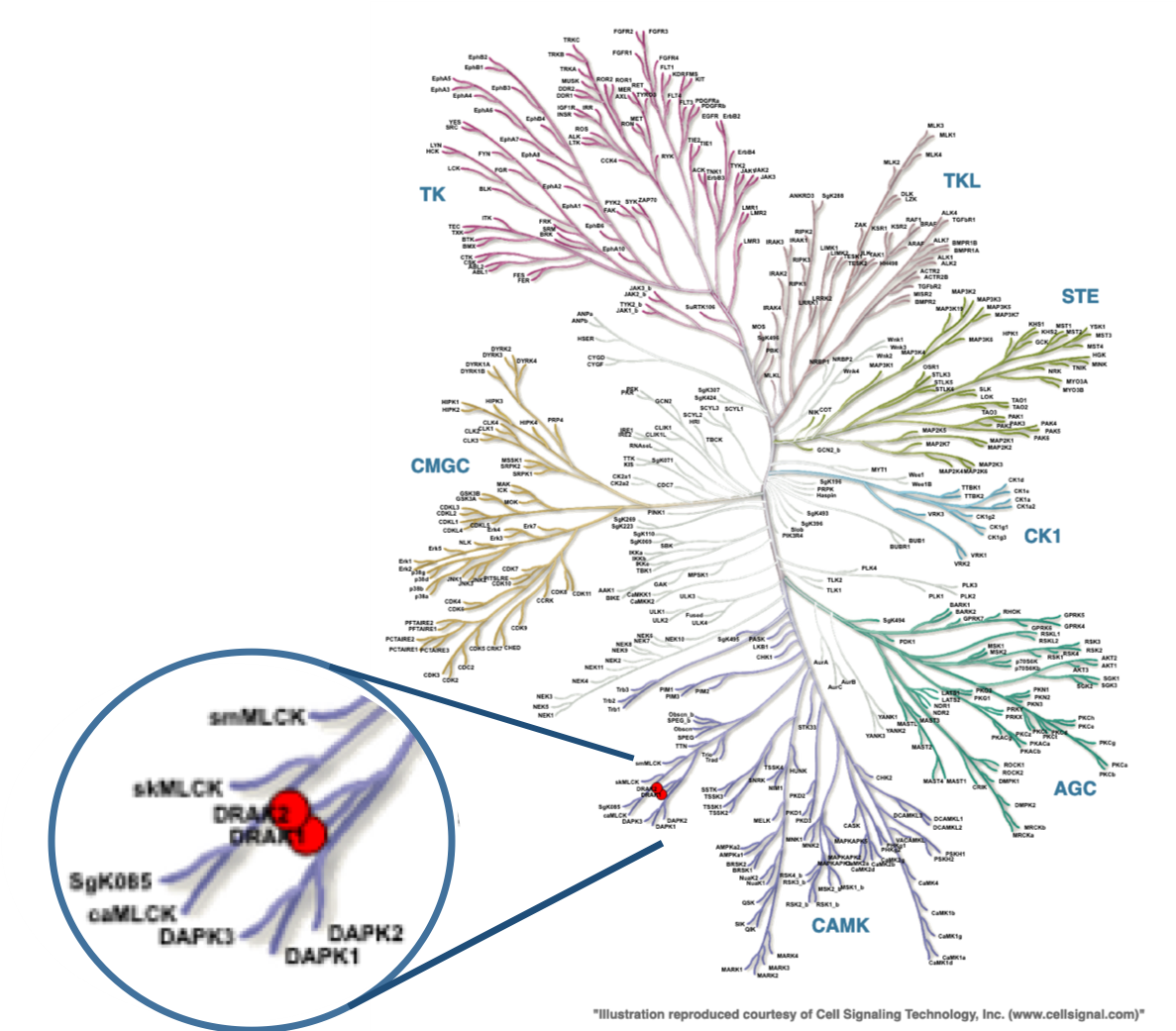
Figure 1. Phylogenetic kinase tree, STK17B/DRAK2 and ST17A/DRAK1 highlighted with red circles. Illustration is reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com).
Selectivity
SGC-STK17B-1 was profiled in the KINOMEscan assay against of 403 wild-type kinases @ 1 mM, followed by Kd determination of potential off-targets at Eurofins. SGC-STK17B-1 shown >30 fold selective (Kd) against the closest off targets STK17A/DRAK1, AURKB and CaMKK2. SGC-STK17B-1 is non-toxic in cells (HEK293 cells) in a time and dose dependent manner and have little to no effect on cell viability.
Cell-based assay
In cellular target engagement assays (NanoBRET) in HEK293 cells, SGC-STK17B-1 showed IC50 of 190 nM against STK17B (n = 9) and >10,000 nM against DRAK1 and AURKB (potential off-targets). The negative control SGC-STK17B-1N shown IC50 > 10,000 nM against STK17B.
Binding Activity
In a binding assay performed at Luceome Biotechnologies SGC-STK17B-1 showed potency of 43 nM against STK17B.
Acknowledgments. NanoBRET cellular assays were performed by Carrow Wells and Julie Pickett at SGC-UNC Chapel Hill.
| Probe |
 |
SGC-STK17B-1 |
| Physical and chemical properties for SGC-STK17B-1 | |
| Molecular weight | 358.46 |
| Molecular formula | C16H10N2O2S3 |
| IUPAC name | 2-((6-(benzo[b]thiophen-2-yl)thieno[3,2-d]pyrimidin-4-yl)thio)acetic acid |
| MollogP | 4.00 |
| PSA | 144.86 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 4 |
| No. of hydrogen bond acceptors | 4 |
| No. of hydrogen bond donors | 1 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10mM) are stable at -20 °C |
| Dissolution | Soluble in DMSO up to 10 mM. |
| Negative control |
 |
SGC-STK17B-1N |
| Physical and chemical properties for SGC-STK17B-1N | |
| Molecular weight | 358.46 |
| Molecular formula | C16H10N2O2S3 |
| IUPAC name | 2-((6-(benzo[b]thiophen-2-yl)thieno[2,3-d]pyrimidin-4-yl)thio)acetic acid |
| MollogP | 4.09 |
| PSA | 144.86 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 4 |
| No. of hydrogen bond acceptors | 4 |
| No. of hydrogen bond donors | 1 |
| Storage | Stable as a solid at room temperature. DMSO stock solutions (up to 10mM) are stable at -20 °C. |
| Dissolution | Soluble in DMSO up to 10 mM. |
SMILES:
SGC-STK17B-1: O=C(CSC1=C2C(C=C(S2)C3=CC4=C(S3)C=CC=C4)=NC=N1)O
SGC-STK17B-1N: O=C(CSC1=C2C(SC(C3=CC4=C(S3)C=CC=C4)=C2)=NC=N1)O
InChI:
SGC-STK17B-1: 1S/C16H10N2O2S3/c19-14(20)7-21-16-15-10(17-8-18-16)6-13(23-15)12-5-9-3-1-2-4-11(9)22-12/h1-6,8H,7H2,(H,19,20)
SGC-STK17B-1N: 1S/C16H10N2O2S3/c19-14(20)7-21-15-10-6-13(23-16(10)18-8-17-15)12-5-9-3-1-2-4-11(9)22-12/h1-6,8H,7H2,(H,19,20)
InChIKey:
SGC-STK17B-1: BNYXRPMABDQJJR-UHFFFAOYSA-N
SGC-STK17B-1N: RRIRDIKTUQVZCW-UHFFFAOYSA-N
The KINOMEscan of SGC-STK17B-1 at 1mM showed few off-targets, Figure 1. With cutoff 10% control (90% inhibition), MET, NEK6, PIM2, WEE1, and CAMKK2 are shown in red circles in the TREEspot. This screening was followed by Kd determination and NanoBRET assays for several potential off-targets (Table 1).
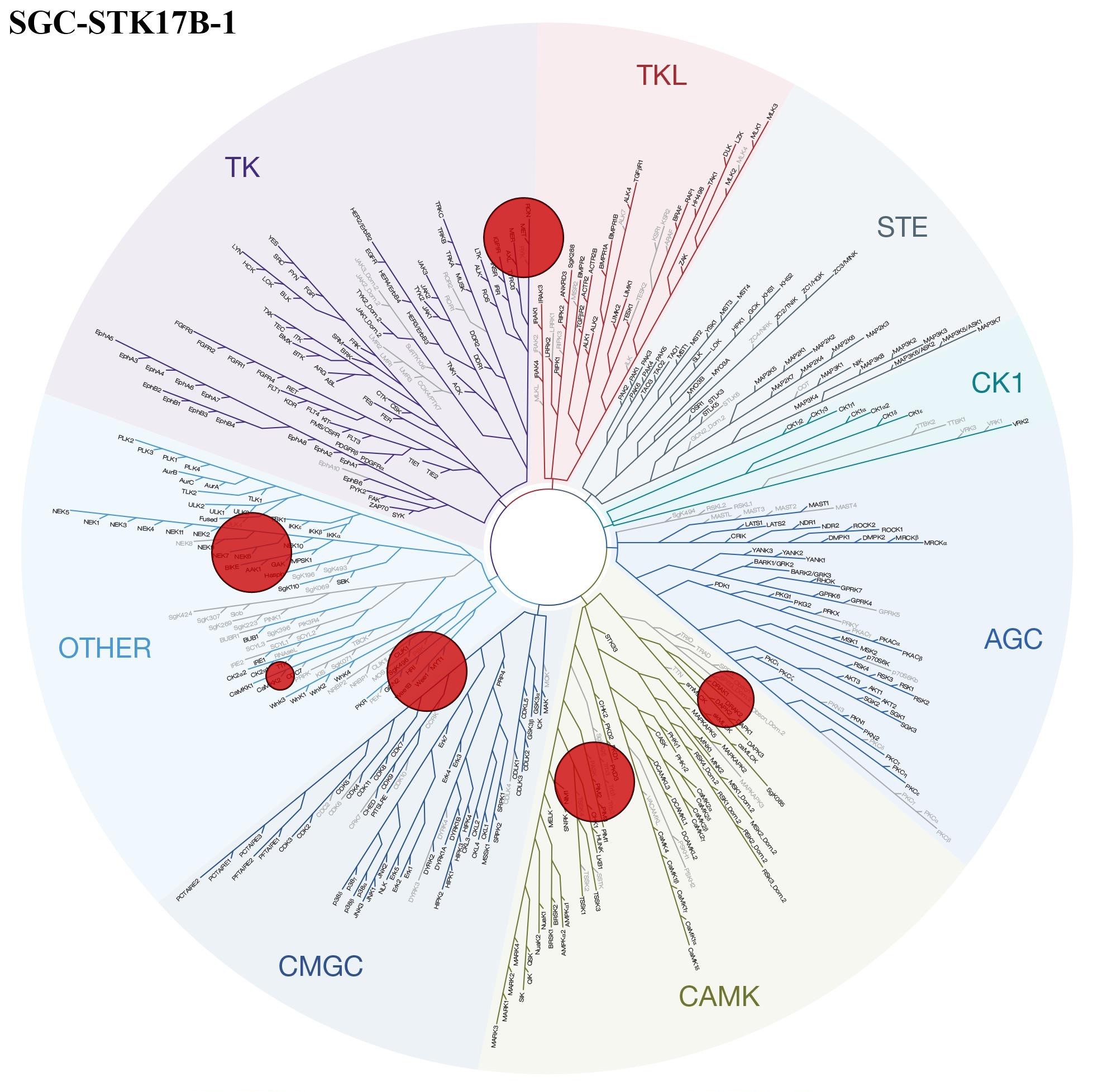
Figure 1: Kinome selectivity and TREEspot analysis of 403 wild-type kinases for SGC-STK17B-1.
Results in Table 1 confirmed the high selectivity of SGC-STK17B-1 towards STK17B/DRAK2. Non-off targets were found for this chemical probe at concentrations shown.
|
|---|
The co-crystal structure shown an unusual flip of R41 (P-loop) forming salt bridge network with inhibitor COOH group, R41 and E117 in one of the two experimentally observed conformations is a likely mechanism of selectivity. PDB depositions for the chemical probe are 6Y6F and 6Y6H (links will be updated as soon as they become available).
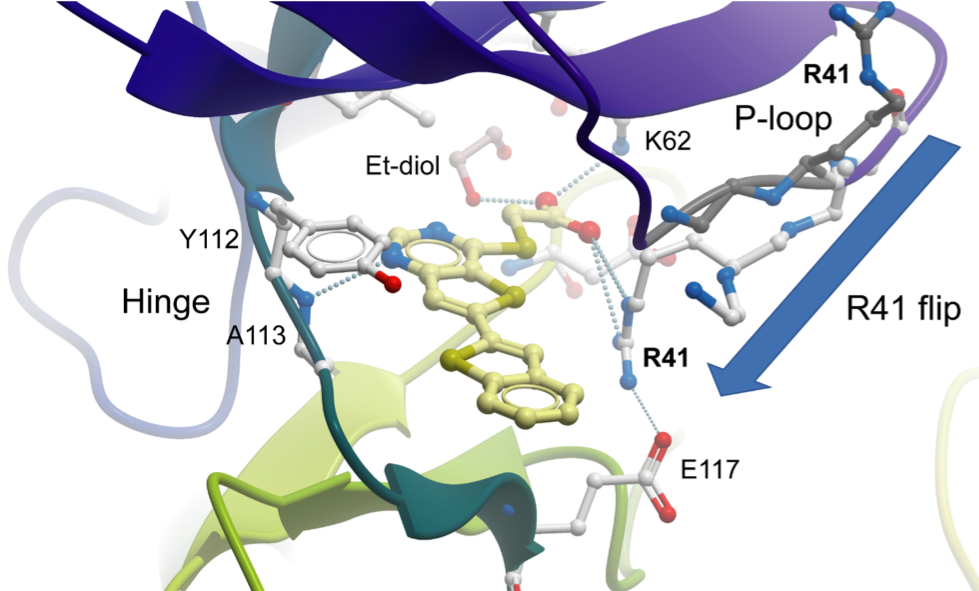
Important features:
Acknowledgments. Co-crystal structure was obtained by Apirat Chaikuad and Stefan Knapp at SGC-Frankfurt.