This probe is available from Tocris and Cayman Chemical.
The control may be requested by clicking here.
| Probe | Negative control | |
 |
|  |
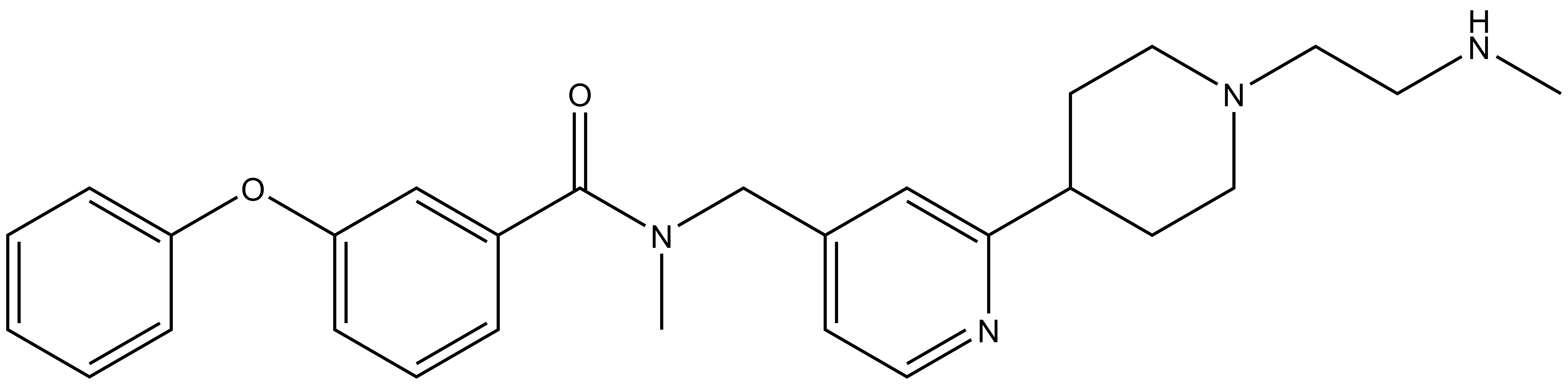
TP-064 |
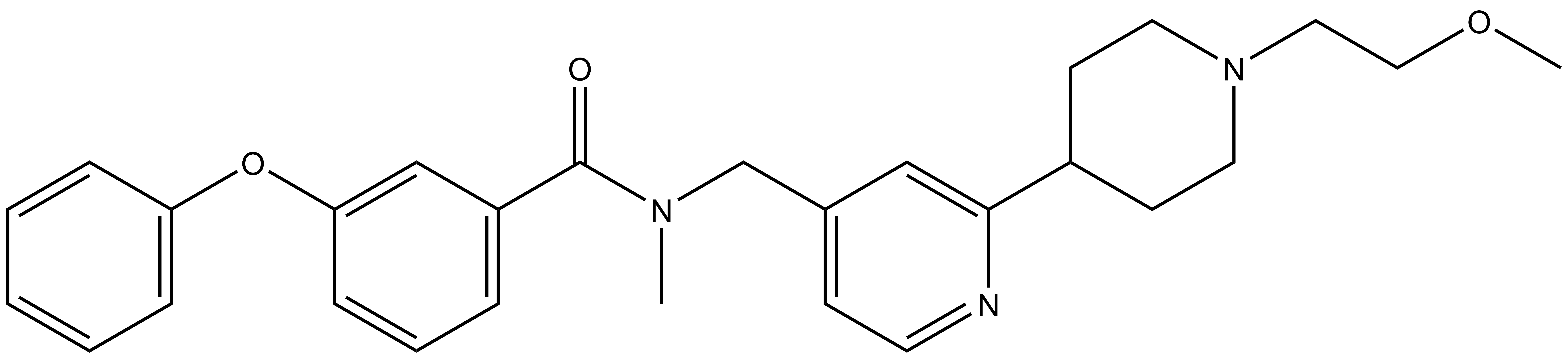
| TP-064N |
A collaboration between Takeda and the SGC has resulted in the discovery of TP-064, the first potent, selective and cell active chemical probe for PRMT4. The in vitro activity of TP-064 includes inhibition of PRMT4 with IC50 < 10nM for methylation of H3 (1-25) and greater than 100-fold selectivity over other histone methyltransferases and non-epigenetic targets. In cellular assays, TP-064 inhibits the methylation of MED12 with IC50 = 43 nM.
A closely related compound, TP-064N, exhibits no activity in the biochemical and cellular assays, and is an ideal control compound for cellular studies.
| Probe | Negative control | |
 |
|  |
TP-064 |
| TP-064N |
| Physical and chemical properties for TP-064 | |
| Molecular weight | 458.3 |
| Molecular formula | C28H34N4O2 |
| IUPAC name | (methyl-((2-(1-(2-methylamino-ethyl)-piperidin-4-yl)-pyridin-4-yl)-methyl)-amino)-(3-phenoxy-phenyl)-methanone |
| MollogP | 3.422 |
| PSA | 47.71 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 10 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 1 |
| Physical and chemical properties for TP-064N (Negative Control) | |
| Molecular weight | 459.3 |
| Molecular formula | C28H33N3O3 |
| IUPAC name | (((2-(1-(2-methoxy-ethyl)-piperidin-4-yl)-pyridin-4-yl)-methyl)-methyl-amino)-(3-phenoxy-phenyl)-methanone |
| MollogP | 4.051 |
| PSA | 44.24 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 10 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 0 |
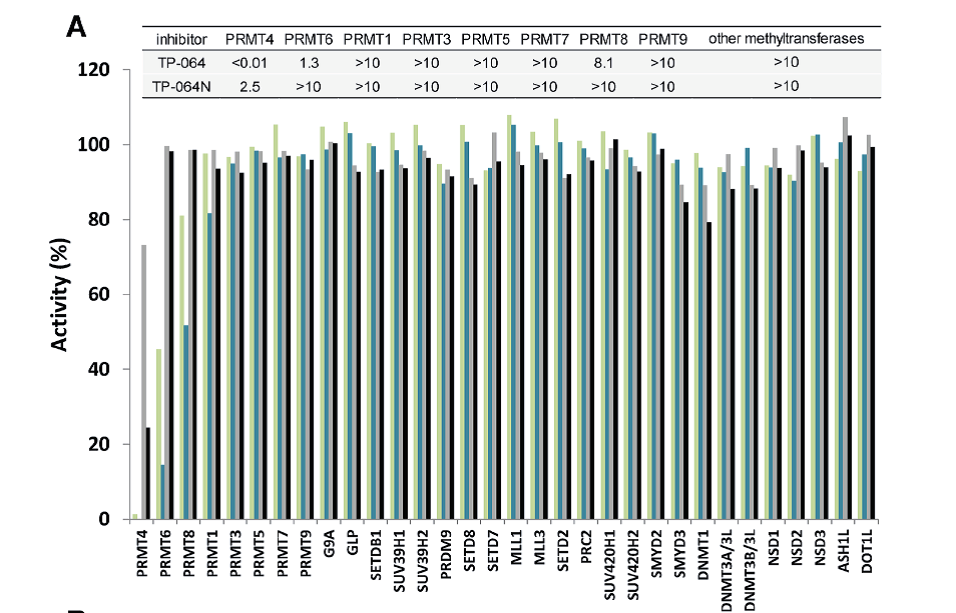
Selectivity of TP-064N at 10 μM () and 1 μM () and of TP-064 at 10 μM () and 1 μM () for PRMT1, 3, 4, 5, 6, 7, 8, and 9 as well as for 24 histone and DNA methyltransferases was assessed. Dose response data are presented in the top panel as IC50s (μM).
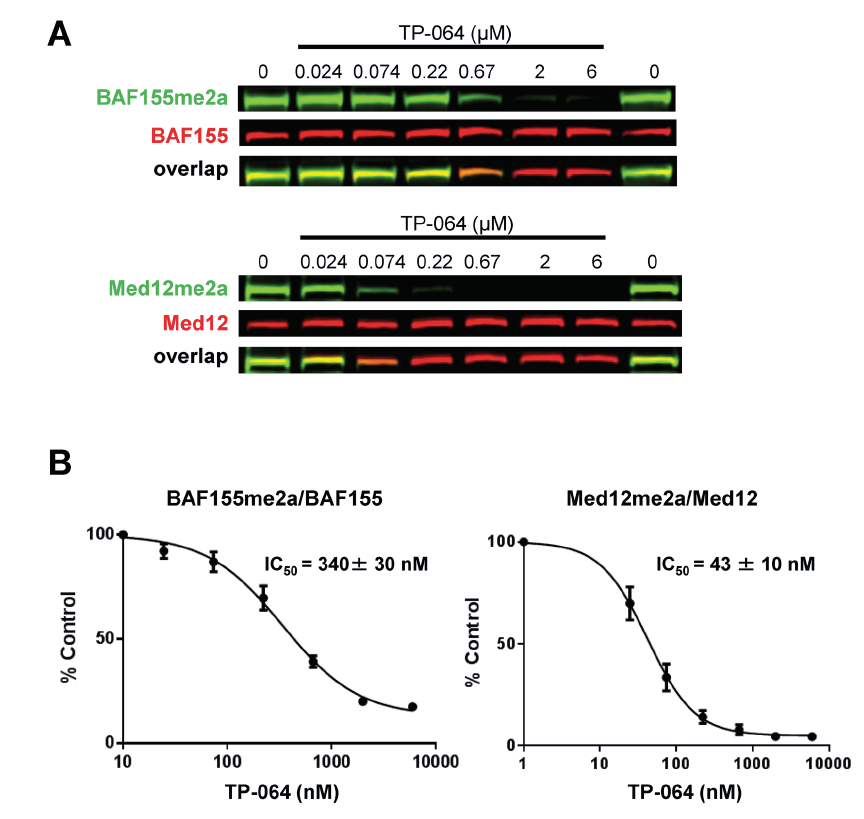
TP-064 inhibits the dimethylation of PRMT4 substrates. HEK293 cells were treated with indicated concentrations of TP-064 for 3 days and dimethylation levels of BAF155 and MED12 in whole cell extracts were analyzed by western blotting. (B) Quantitation of data in (A). Graphs represent nonlinear curve fits of dimethyl-BAF155 and dimethyl-MED12 signal intensities normalized to total BAF155 or MED12, respectively. Data represent mean ± SEM of two independent experiments prepared in triplicate.
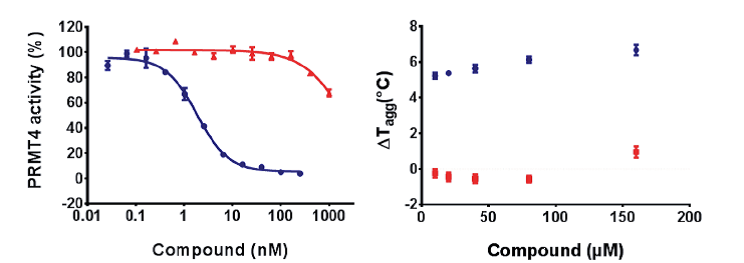
TP-064 (blue) inhibits PRMT4 activity with an IC50 value of < 10 nM under balanced conditions. TP-064N (red) has no effect on PRMT4 activity up to 100 nM. The binding of TP-064 to PRMT4 was confirmed by DSLS with stabilization at about 6°C. No binding was observed with TP-064N
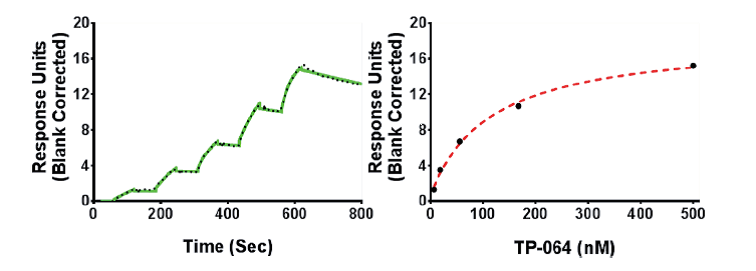
SPR analysis of the TP-064 binding to PRMT4 in the presence of 50 μM SAM. A representative sensorgram (black dots) is shown with the kinetic fit (solid green). A Kd value of 7.1 ± 1.8 nM, with kon = 1.1 ± 0.1 × 105 M−1 s−1 and koff = 0.7 ± 0.1 × 10−3 s−1, was obtained from triplicate experiments. The steady state response (black circles) and 1:1 binding model fitting (red dashed line) is presented.
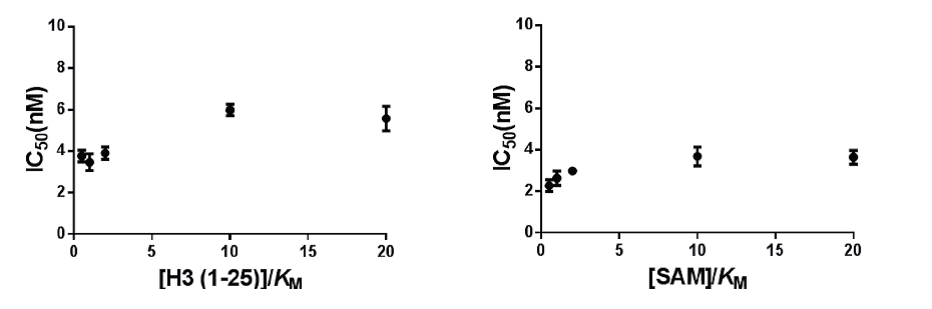
Mechanism of action of TP-064 was assessed by determining IC50 of both substrates values at various concentrations.
Kazuhide Nakayama, Magdalena M. Szewczyk, Carlo dela Sena, Hong Wu, Aiping Dong, Hong Zeng, Fengling Li, Renato Ferreira de Freitas, Mohammad S. Eram, Matthieu Schapira, Yuji Baba, Mihoko Kunitomo, Douglas R. Cary, Michiko Tawada, Akihiro Ohashi, Yasuhiro Imaeda, Kumar Singh Saikatendu, Charles E. Grimshaw, Masoud Vedadi, Cheryl H. Arrowsmith, Dalia Barsyte-Lovejoy, Atsushi Kiba, Daisuke Tomita and Peter J. Brown TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget 9: 18480-93 (2018)

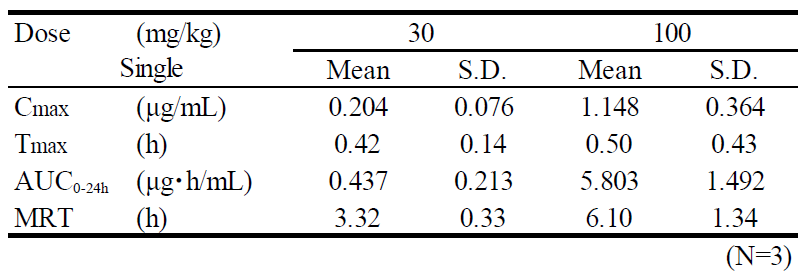
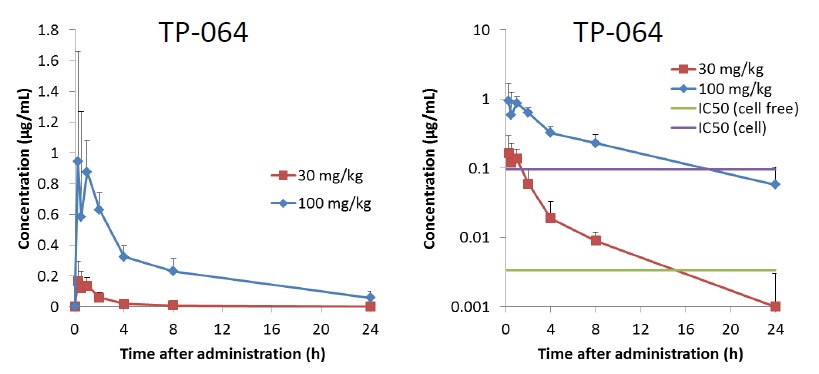
| Test compound | TP-064 |
| Animal | C.B-17/Icr-scid mouse, female, 7W, fed |
| Formulation | 10% DMSO/10% Cremophor EL /10% PropyleneGlycol (PG)/Distilled Water |
| Administration | i.p., 30 or 100 mg/kg as free body |
| Test items | Plasma |
| Sampling time | 0.25, 0.5, 1, 2, 4, 8, 24 hours after administration |
Main features