This probe is available from Tocris, Cayman Chemical and Sigma.
Its negative control (TP-472N) is available for purchase from Tocris and Sigma.
| Probe | Negative control | |
 |
|  |
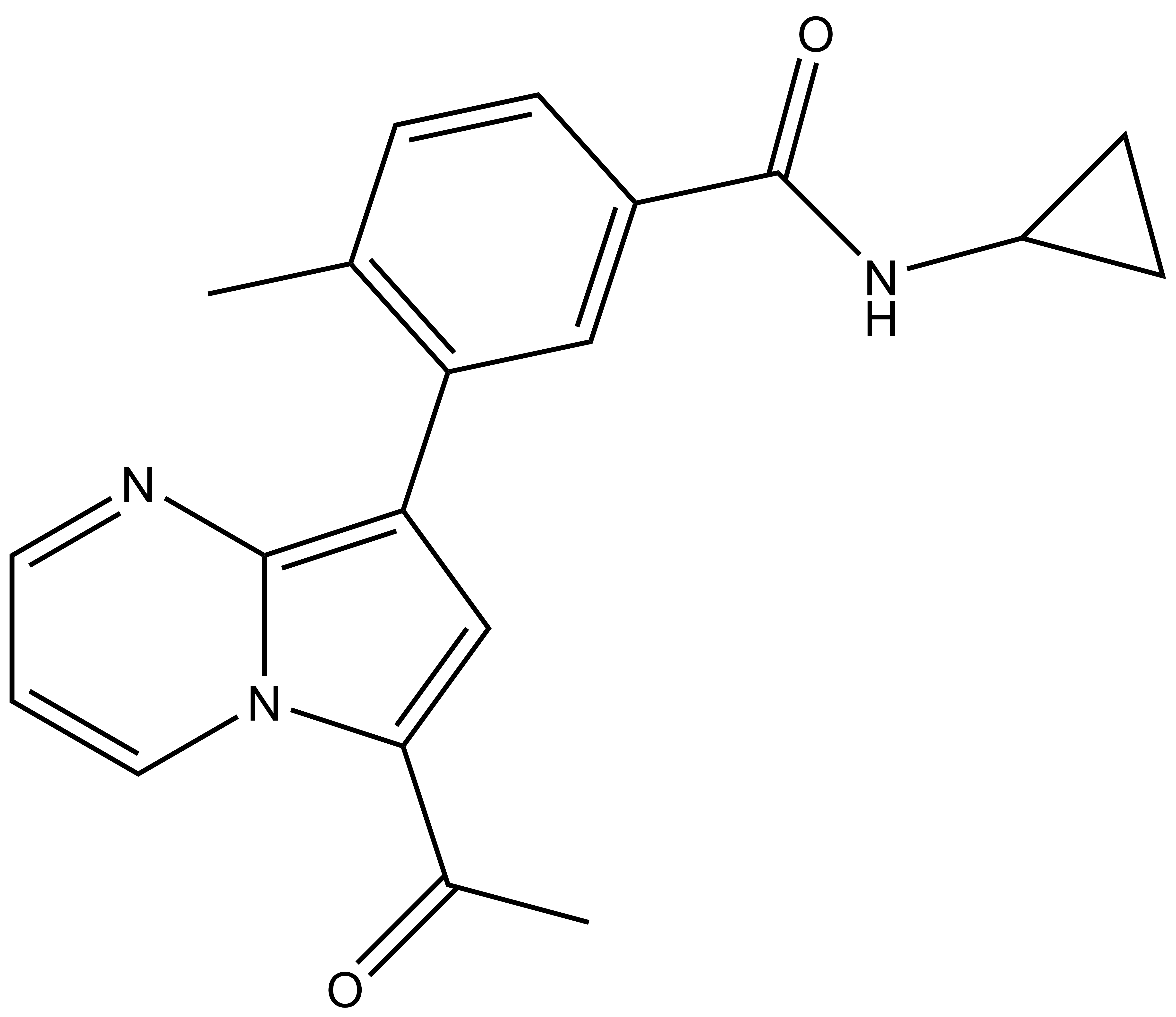
TP-472 |
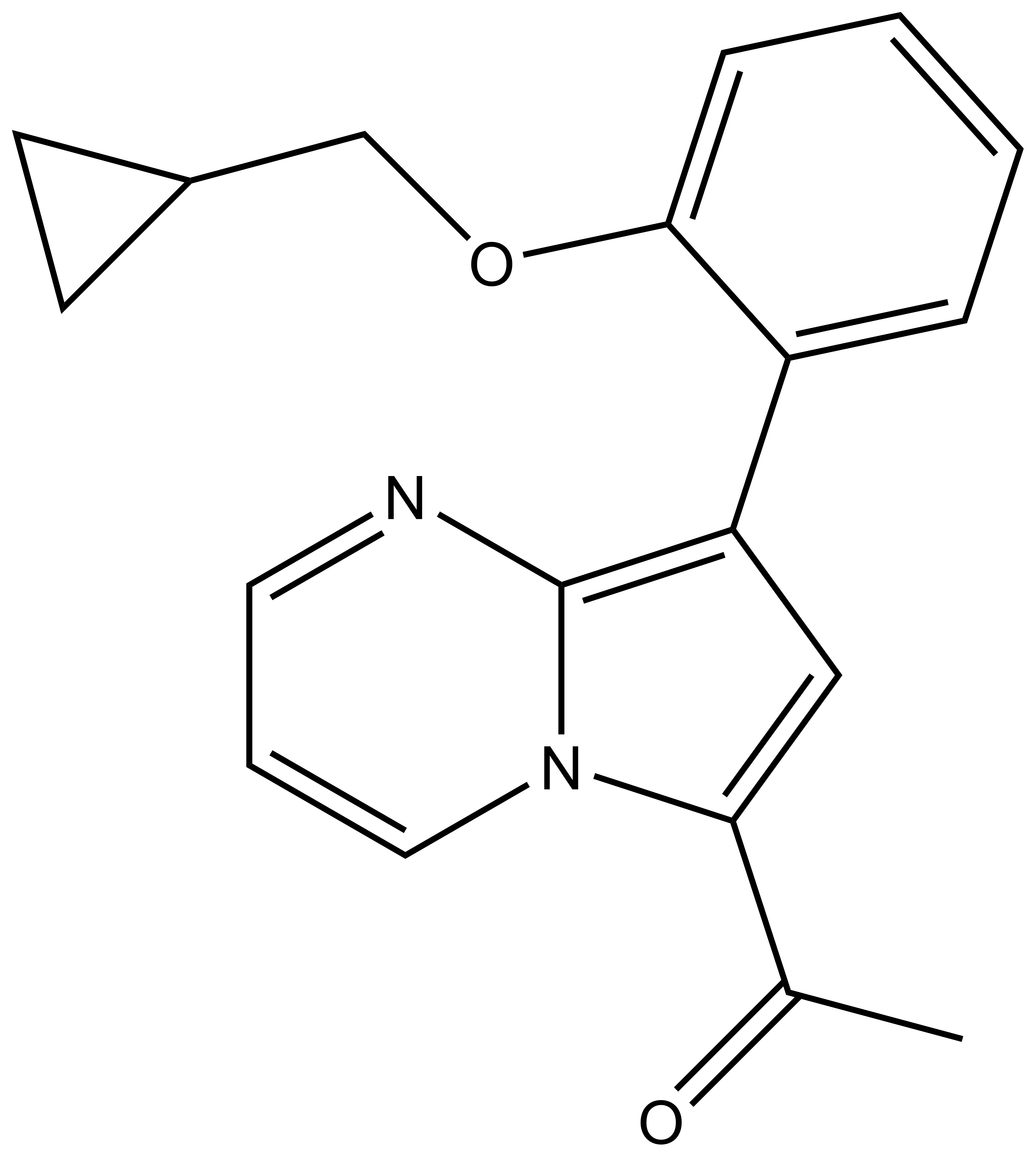
| TP-472N |
The bromodomain containing protein 9 (BRD9) has been reported as a component of the switch/sucrose non-fermentable (SWI/SNF) brahma-related gene 1-associated factor (BAF) complex, which plays a key role in chromatin remodelling and transcription control [1] although the precise biological role is unknown. The highly homologous bromodomain containing protein 7 (BRD7) is a component of the SWI/SNF polybromo-associated BAF (PBAF) complex and has been proposed as a tumor suppressor [2].
In a collaborative effort Takeda and the SGC have identified and characterised TP-472 as a BRD9/7 probe. This molecule is of a different chemotype to the BRD9/7 probes previously available, has a structurally similar negative control available (TP-472N), has a good PK profile and is suitable for in vivo applications and is synthetically tractable.
TP-472 has excellent potency (BRD9 KD 33nM; BRD7 340nM by ITC), selectivity >30 fold over all other bromodomain family members except BRD7 and is cell active (EC50 320 nM in a BRD9 NanoBRET assay). The negative control TP472N is inactive against other bromodomains (>20uM against BRD9)
| Probe | Negative control | |
 |
|  |
TP-472 |
| TP-472N |
| Physical and chemical properties for TP-472 | |
| Molecular weight | 333.1 |
| Molecular formula | C20H19N3O2 |
| IUPAC name | 1-(7-(5-(cyclopropylamino-formyl)-2-methyl-phenyl)-1,5-diaza-bicyclo[4.3.0]nona-2,4,6,8-tetraen-9-yl)-ethanone |
| MollogP | 3.102 |
| PSA | 46.85 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 5 |
| No. of hydrogen bond acceptors | 5 |
| No. of hydrogen bond donors | 1 |
| Physical and chemical properties for TP-472N (Negative Control) | |
| Molecular weight | 306.1 |
| Molecular formula | C19H18N2O2 |
| IUPAC name | 1-(7-(2-(cyclopropyl-methoxy)-phenyl)-1,5-diaza-bicyclo[4.3.0]nona-2,4,6,8-tetraen-9-yl)-ethanone |
| MollogP | 3.673 |
| PSA | 30.22 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 5 |
| No. of hydrogen bond acceptors | 4 |
| No. of hydrogen bond donors | 0 |
Selectivity beyond target family
TP-472 and TP-472N were screened in the Eurofins CEREP Diversity Profile to measure binding to a range of Receptors, Ion Channels and enzymes @10 uM. The Inhibition as the mean % of control is shown below, results showing an inhibition or stimulation lower than 50% are considered to represent significant effects. TP-472 showed binding to Adenosine A1 receptor (14%), Benzodiazepine receptor (47%), PDE2A1 (h) (25%), PDE3A (h) (48%) and PDE4D2 (h) (28%). TP-472N showed binding to A1 receptor (35%), A3 receptor (23%), Melatonin receptor MT1 (47%) and Cl- channel (GABA-gated) (20%).