| Probe | Negative control | |
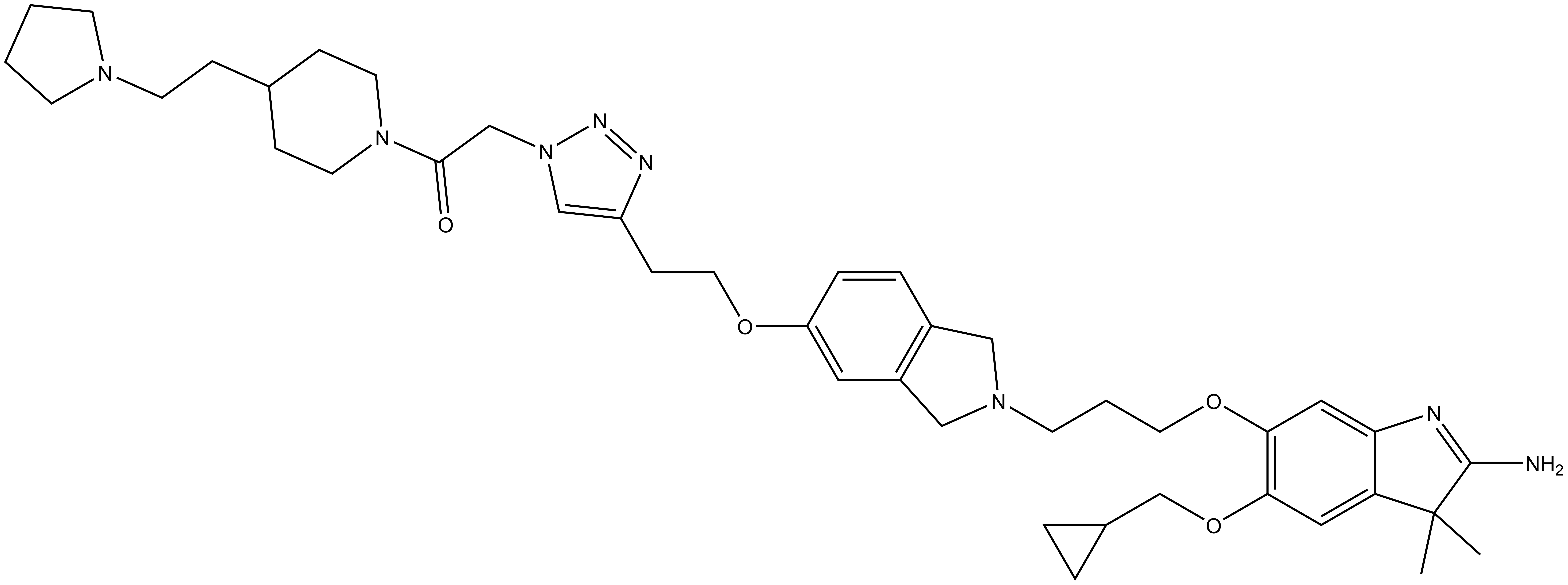 | 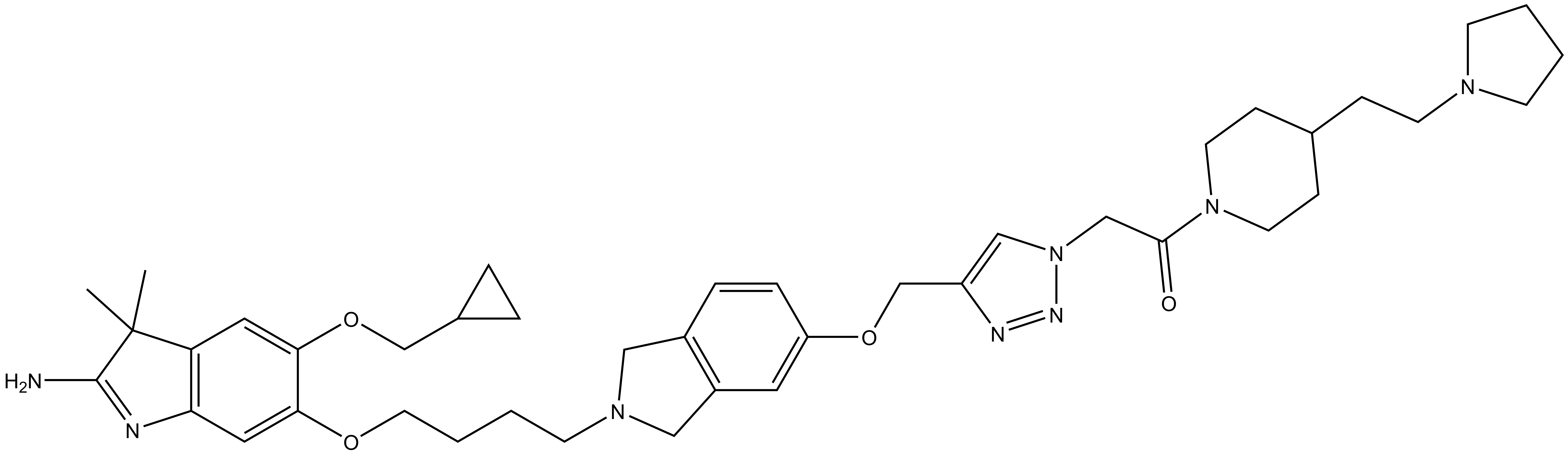 | |
VinSpinIn (Vinnie's Spindlin Inhibitor) | VinSpinIC (Vinnie's Spindlin Inactive Control) |
The Spindlin proteins are tudor domain containing proteins. There are five spindlin family members (Spin1, Spin2A, Spin2B, Spin3, Spin4), which are expressed at various levels throughout the body. However, spindlin1 (Spin1) is expressed at higher levels compared to the other family members.1 Our current knowledge of the biological roles of spindlins is limited to that of Spin1.
The Spin proteins consist of three tudor-like Spin/Ssty domains, arranged in a clockwise orientation.2-4 Spin1 has been shown to bind to trimethylated lysine 4 of histone 3 (H3K4me3) via domain 2 (ITC Kd = 147 nM).3,5 However, increased binding is observed when a second epigenetic methylation mark is present on arginine 8 (asymmetrically dimethylated) (H3K4me3R8me2a; ITC Kd = 45 nM).2 Spin1 also binds to H4K20me3 via domain 2.6
Spin1 binding to methylated histones is associated with transcriptional activation. Spin1 was found to be overexpressed in various cancers and has been shown to drive cancer cell proliferation through activation of the Wnt/β-catenin, PI3K/Akt and RET signalling pathways.2,7-10
Conversely, Spin1 has been shown to facilitate the inactivation of p53 by sequestering the ribosomal protein uL18.11
Direct or indirect Spin1 knockdown resulted in cancer cell and xenograft tumor growth inhibition, and such studies suggest that small molecule inhibition of Spin1 may be a viable approach for the treatment of certain cancers.8-13
Therefore, the SGC has developed VinSpinIn as a potent, cell active chemical probe for the Spin family proteins. VinSpinIn, along with the structurally very similar inactive control compound VinSpinIC, will contribute significantly to the elucidation of the biological roles and functions of the spindlin proteins, and will aid with the validation of Spin1 as a chemotherapeutic target.
Potency Against Target Family
The SYPRO Orange thermal shift assay was employed to assess the potency of VinSpinIn and VinSpinIC against four of the five Spin family members (Table 1). VinSpinIn induced a large shift in the thermal stability in all of the Spin protein assessed, while VinSpinIC did not. ITC was also performed to determine the potency of VinSpinIn on four Spin family members as well as an additional Spin1 construct (Table 1). VinSpinIn had KDs ranging between approximately 10-130 nM across the family.
| Spin Family Proteins | Thermal Shift Assay (ΔTm°C) | ITC Assay (KD nM) | |
| VinSpinIn | VinSpinIC | VinSpinIn | |
| Spin149-262 | Not tested | Not tested | 9.9 |
| SPIN126-262 | 13.17 | 1.02 | 111.1 |
| SPIN2B22-258 | 10.47 | 0.29 | 46.1 |
| SPIN321-258 | 14.12 | 2.34 | 131.1 |
| SPIN436-249 | 6.53 | 0.25 | 18.1 |
Table 1: Potency Against Target Family
Selectivity
VinSpinIn and VinSpinIC were screened against a panel of methyl binding domains (MBDs) using the thermal shift assay (Table 2). No significant thermal shift was observed for any of the MBDs screened.
A Scintillation Proximity Assay (SPA) was employed to screen VinSpinIn and VinSpinIC against a panel of methyltransferases and IC50s were determine for selected targets (Table 3). The lowest IC50 of VinSpinIn (PRMT4) was approximately 300 times greater than the AlphaScreen IC50 on Spin1 (30 nM).
| Methyl Lysine Binders | VinSpinIn (ΔTm°C) | VinSpinIC (ΔTm°C) |
| UHRF1139-298 | -0.04 | -0.14 |
| 53BP11483-1606 | -0.65 | -0.41 |
| TDRD3525-611 | 0.15 | -0.95 |
| SND1650-910 | -0.09 | -1.62 |
| SETDB1197-403 | -0.04 | -0.34 |
| SGF29129-293 | -0.05 | -0.92 |
| CCDC101114-293 | -0.04 | 0.3 |
| CHD1269-446 | -0.65 | 2.37 |
| FALZ2736-2793 | 0.15 | 0.31 |
| ING2211-266 | -0.09 | 0.25 |
| JARID1A1542-1660 | -0.04 | -0.04 |
| MLL1558-1773 | -0.05 | 0.63 |
| MLL5113-171 | -0.05 | -0.05 |
| PHF21-64 | 0.02 | 0.19 |
| PHF81-63 | 0.23 | -0.11 |
| TAF31086-1153 | -0.21 | -0.04 |
Table 2: Screening on a panel of MBDs.
| Methyltransferase Panel | VinSpinIn (IC50 µM) | VinSpinIC (IC50 µM) |
| PRMT4 | 9 | 2 |
| SETD2 | 20 | 5 |
| PRMT7 | 19 | 8 |
| SUV39H1 | 34 | 18 |
| PRMT6 | 27 | 19 |
| PRC2 | 21 | 27 |
| SMYD2 | 22 | 44 |
| PRDM9 | 25 | 44 |
| PRMT1 | 47 | 59 |
| PRMT8 | 45 | 66 |
Table 3: IC50s on methyltransferases.
Dosage
Use between 0.5 and 3 µM for Cellular Assays and 1 µM for screening at a single shot, for both VinSpinIn & VinSpinIC.
In vitro Activity
In a number of biophysical assays, VinSpinIn was shown to be a potent Spin1 inhibitor and has an ITC KD which is approximately 130 times more potent than that of the inactive control VinSpinIC (Table 4).
| Compound | AlphaScreen (Spin126-262) (IC50) | Octet BLI (Spin126-262) (KD) | ITC (Spin126-262) (KD) | ITC (Spin149-262) (KD) | Tm Shift (Spin126-262) (ΔTm°C) |
| VinSpinIn | 30 nM | 55 nM | 111.1 nM | 9.9 nM | 13.2 |
| VinSpinIC | 3.64 µM | not tested | not tested | 1.3 µM | 1.0 |
Table 4: In vitro biophysical binding assay results of VinSpinIn & VinSpinIC.
Cellular Activity
In a NanoBRET cellular target engagement assay VinSpinIn displayed dose dependant inhibition of the Spin1-H3 interaction; the inactive VinSpinIC showed no inhibition (Figure 2).
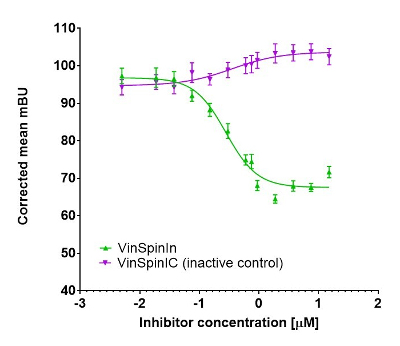
Figure 2: NanoBRET cellular engagement assay. Click on the ‘Cell-based Assay Data’ tab above for more details.
| VinSpinIn |
 |
Click here to download the SDF file. |
| VinSpinIn | |
| Physical and chemical properties | |
| Molecular weight | 738.96 g/mol |
| Molecular formula | C42H58N8O4 |
| IUPAC name | 2-[4-(2-{[2-(3-{[2-amino-5-(cyclopropylmethoxy)-3,3-dimethyl-3H-indol-6-yl]oxy}propyl)-2,3-dihydro-1H-isoindol-5-yl]oxy}ethyl)-1H-1,2,3-triazol-1-yl]-1-{4-[2-(pyrrolidin-1-yl)ethyl]piperidin-1-yl}ethan-1-one |
| logP | 3.85 |
| TPSA | 123.57 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 17 |
| No. of hydrogen bond acceptors | 2 |
| No. of hydrogen bond donors | 12 |
| Storage | +4 °C |
| Dissolution | DMSO (up to at least 50 mM) |
| VinSpinIC |
 |
Click here to download the SDF file. |
| VinSpinIC | |
| Physical and chemical properties | |
| Molecular weight | 738.96 g/mol |
| Molecular formula | C42H58N8O4 |
| IUPAC name | 2-[4-({[2-(4-{[2-amino-5-(cyclopropylmethoxy)-3,3-dimethyl-3H-indol-6-yl]oxy}butyl)-2,3-dihydro-1H-isoindol-5-yl]oxy}methyl)-1H-1,2,3-triazol-1-yl]-1-{4-[2-(pyrrolidin-1-yl)ethyl]piperidin-1-yl}ethan-1-one |
| clogP | 3.99 |
| TPSA | 123.57 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 17 |
| No. of hydrogen bond acceptors | 2 |
| No. of hydrogen bond donors | 12 |
| Storage | +4 °C |
| Dissolution | DMSO (up to at least 50 mM) |
SMILES:
VinSpinIn: CC1(C(N)=NC2=CC(OCCCN3CC4=C(C3)C=C(OCCC5=CN(N=N5)CC(N6CCC(CC6)CCN7CCCC7)=O)C=C4)=C(C=C21)OCC8CC8)C
VinSpinIC: O=C(CN1N=NC(COC2=CC3=C(C=C2)CN(C3)CCCCOC4=CC5=C(C(C)(C(N)=N5)C)C=C4OCC6CC6)=C1)N7CCC(CC7)CCN8CCCC8
InChI:
VinSpinIn: InChI=1/C42H58N8O4/c1-42(2)36-23-38(54-29-31-6-7-31)39(24-37(36)44-41(42)43)53-20-5-16-48-25-32-8-9-35(22-33(32)26-48)52-21-13-34-27-50(46-45-34)28-40(51)49-18-11-30(12-19-49)10-17-47-14-3-4-15-47/h8-9,22-24,27,30-31H,3-7,10-21,25-26,28-29H2,1-2H3,(H2,43,44)/f/h43H2
VinSpinIC: InChI=1/C42H58N8O4/c1-42(2)36-22-38(54-28-31-7-8-31)39(23-37(36)44-41(42)43)52-20-6-5-16-48-24-32-9-10-35(21-33(32)25-48)53-29-34-26-50(46-45-34)27-40(51)49-18-12-30(13-19-49)11-17-47-14-3-4-15-47/h9-10,21-23,26,30-31H,3-8,11-20,24-25,27-29H2,1-2H3,(H2,43,44)/f/h43H2
InChIKey:
VinSpinIn: XPEJZXWPKDAYFX-UHFFFAOYSA-N
VinSpinIC: FOBGCBSESHUBEI-UHFFFAOYSA-N
The SYPRO Orange thermal shift assay was employed to screen VinSpinIn and VinSpinIC against a panel of methyl binding domains (MBDs), which included four of the five Spin family members (Table 1). VinSpinIn induced a large shift in thermal stability of Spin1, Spin2B, Spin3, Spin4. No other significant shift in thermal stability was observed for VinSpinIn or the inactive VinSpinIC.
| Methyl Lysine Binders | VinSpinIn (ΔTm°C) | VinSpinIC (ΔTm°C) |
| a UHRF1139-298 | -0.04 | -0.14 |
| a 53BP11483-1606 | -0.65 | -0.41 |
| a TDRD3525-611 | 0.15 | -0.95 |
| a SND1650-910 | -0.09 | -1.62 |
| a SETDB1197-403 | -0.04 | -0.34 |
| a SGF29129-293 | -0.05 | -0.92 |
| b CCDC101114-293 | -0.04 | 0.3 |
| b CHD1269-446 | -0.65 | 2.37 |
| b FALZ2736-2793 | 0.15 | 0.31 |
| b ING2211-266 | -0.09 | 0.25 |
| b JARID1A1542-1660 | -0.04 | -0.04 |
| b MLL1558-1773 | -0.05 | 0.63 |
| b MLL5113-171 | -0.05 | -0.05 |
| b PHF21-64 | 0.02 | 0.19 |
| b PHF81-63 | 0.23 | -0.11 |
| b TAF31086-1153 | -0.21 | -0.04 |
| b SPIN126-262 | 13.17 | 1.02 |
| b SPIN2B22-258 | 10.47 | 0.29 |
| b,c SPIN321-258 | 14.12 | 2.34 |
| b SPIN436-249 | 6.53 | 0.25 |
Table 1: Screening on a panel of MBDs.a protein = 0.05 to 0.2 mg/ml, compound = 200 µM; b protein = 2 µM, compound = 20 µM; c Intrinsic tryptophan fluorescence used
A fluorescence polarization displacement assays was employed to screen the compounds against L3MBTL1 and L3MBTL3 (MBT methyl lysine readers), which gave respective Kdisp.s of 27 and 8 µM for VinSpinIn, and 28 and 14 µM for the inactive VinSpinIC. Therefore, VinSpinIn is approximately 267 times less potent towards L3MBTL1 than Spin1.
Selectivity screening was also performed against a panel of methyl transferases domains (MTDs) including protein, DNA and RNA methyltransferases using a Scintillation Proximity Assay (SPA) (Figure 1).
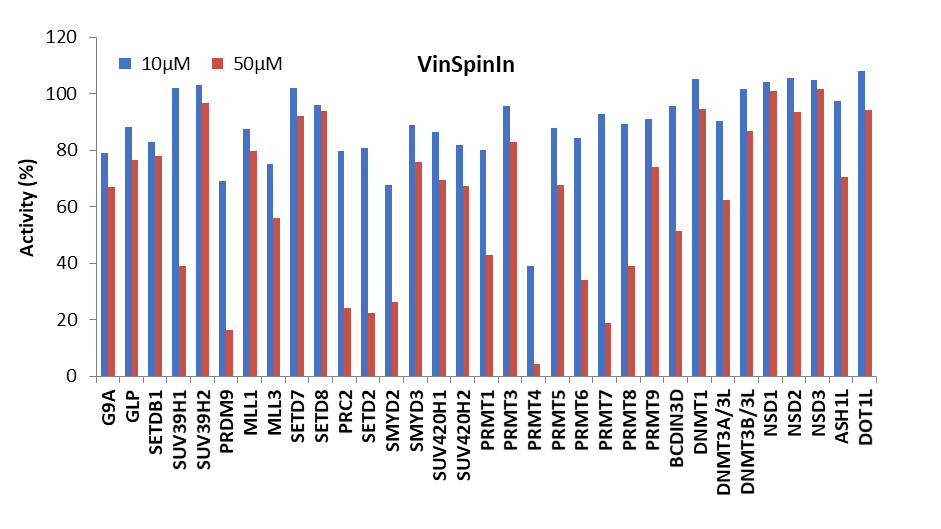
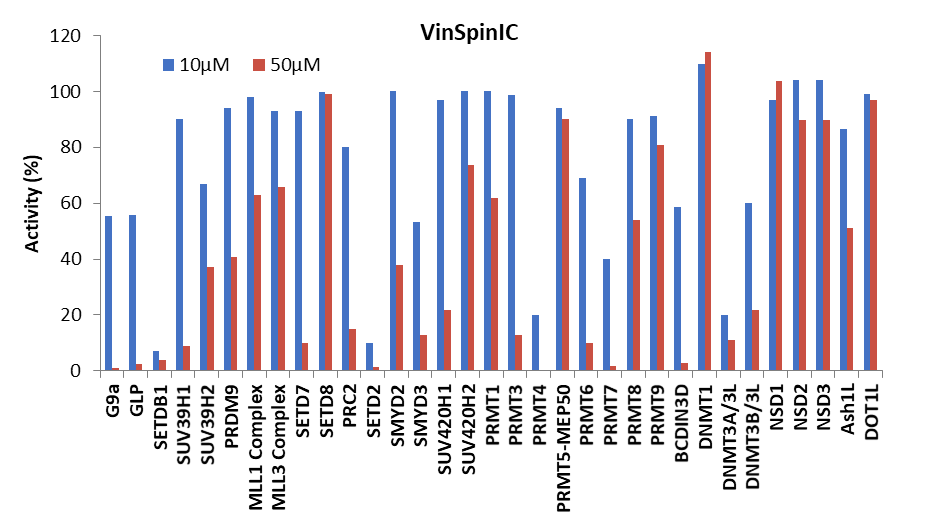
Figure 1: (Top) Methyltransferase activity in presence of VinSpinIn at 10 & 50 µM
(Bottom) Methyltransferase activity in presence of VinSpinIC at 10 and 50 µM.
For selected targets IC50s were determined (Table 2). The lowest IC50 of VinSpinIn (PRMT4) was approximately 300 times greater than the AlphaScreen IC50 on Spin1.
| Methyltransferase Panel | VinSpinIn (IC50 µM) | VinSpinIC (IC50 µM) |
| PRMT4 | 9 | 2 |
| SETD2 | 20 | 5 |
| PRMT7 | 19 | 8 |
| SUV39H1 | 34 | 18 |
| PRMT6 | 27 | 19 |
| PRC2 | 21 | 27 |
| SMYD2 | 22 | 44 |
| PRDM9 | 25 | 44 |
| PRMT1 | 47 | 59 |
| PRMT8 | 45 | 66 |
| DNMT3A/3L | a | 9 |
| DNMT3B/3L | a | 5 |
| G9a | a | 9 |
| GLP | a | 21 |
| SETDB1 | a | 6 |
Table 2: IC50s of selected methyltransferases.a Not determined
Effects of VinSpinIn and VinSpinIC on methyltransferase activity of G9a, GLP, SUV39H1, SUV39H2, SETDB1, SETD8, SUV420H1, SUV420H2, SETD7, MLL1 trimeric complex, MLL3 pentameric complex, EZH2 trimeric complex, PRMT1, PRMT3, PRMT4, PRMT5-MEP50 complex, PRMT6, PRMT7, PRMT8, PRMT9, PRDM9, SETD2, SMYD2, SMYD3, and DNMT1 was assessed by monitoring the incorporation of tritium-labeled methyl group to lysine or arginine residues of peptide substrates using Scintillation Proximity Assay (SPA) as previously described14. Assays were performed in a 10 µl reaction mixture containing 3H-SAM (Cat.# NET155V250UC; Perkin Elmer; www.perkinelmer.com) at substrate concentrations close to Km values for each enzyme. Two concentrations (10µM and 50 µM) of VinSpinIn or VinSpinIC were used in all selectivity assays. To stop the enzymatic reactions, 10 µl of 7.5 M guanidine hydrochloride was added, followed by 180 µl of buffer (20 mM Tris, pH 8.0), mixed and then transferred to a FlashPlate (Cat.# SMP103; Perkin Elmer; www.perkinelmer.com). After mixing, the reaction mixtures in Flash plates were incubated for 2 hours and the CPM were measured using Topcount plate reader (Perkin Elmer, www.perkinelmer.com). The CPM counts in the absence of compound for each data set were defined as 100% activity. In the absence of the enzyme, the CPM counts in each data set were defined as background (0%).
For DOT1L, NSD1, NSD2, NSD3, ASH1L, DNMT3A/3L, and DNMT3B/3L, a filter-based assay was used. In this assay, 10 µl of reaction mixtures were incubated at 23 oC for 1 hour, 50 µl of 10% trichloroacetic acid (TCA) was added, mixed and transferred to filter-plates (Millipore; cat.# MSFBN6B10; www.millipore.com). Plates were centrifuged at 2000 rpm (Allegra X-15R - Beckman Coulter, Inc.) for 2 min followed by 2 additional 10% TCA wash and one ethanol wash followed by centrifugation. Plates were dried and 30 µl MicroO (MicroScint-O; Cat.# 6013611, Perkin Elmer; www.perkinelmer.com) was added to each well, centrifuged and removed. 50 µl of MicroO was added again and CPM was measured using Topcount plate reader.
IC50 Determinations:
IC50 values were determined for inhibition of methyltransferase activity of the following enzymes:
To stop the enzymatic reactions, 7.5 M Guanidine hydrochloride was added, followed by 180 µL of buffer (20 mM Tris, pH 8.0), mixed and then transferred to a FlashPlate (Cat.# SMP103; Perkin Elmer; www.perkinelmer.com). After mixing, the reaction mixtures in Flash plate were incubated for 2 hour and the CPM counts were measured using Topcount plate reader (Perkin Elmer, www.perkinelmer.com). The CPM counts in the absence of compound for each data set were defined as 100% activity. In the absence of the enzyme, the CPM counts in each data set were defined as background (0%). The IC50 values were calculated using GraphPad Prism 7 software.
In a NanoBRET cellular target engagement assay VinSpinIn displayed dose dependant inhibition of the Spin1-H3 interaction, with an IC50 of 270 nM. The inactive VinSpinIC showed no inhibition (Figure 1).
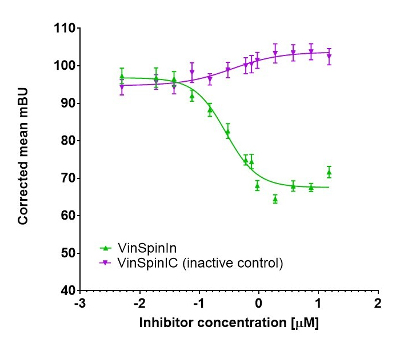
Figure 1: NanoBRET cellular target engagement assay performed in U2OS cells. Full length SPIN1 with N-terminal NanoLuc; Histone 3.3 with C-terminal Halotag; 24h incubation with VinSpinIn and VinSpinIC
U20S cell (2.8 x 105) were plated in each well of a 6-well plate after 6h cells were co-transfected with C-terminal HaloTag-Histone 3.3 (NM_002107) and an N-terminal NanoLuciferase fusion of full length SPIN1 at a 1:500 (NanoLuc® to HaloTag®) ratio respectively with FuGENE HD transfection regent15.
Sixteen hours post-transfection, cells were collected, washed with PBS, and exchanged into media containing phenol red-free DMEM and 4% FBS in the absence (control sample) or the presence (experimental sample) of 100 nM NanoBRET 618 fluorescent ligand (Promega). Cells were then re-plated in a 384-well assay white plate (Greiner #3570) at 2.7x103 cells per well. VinSpinIn and VinSpinIC were then added directly to media at final concentrations 0-30μM or an equivalent amount of DMSO as a vehicle control, and the plates were incubated for 24 h at 37oC in the presence of 5% CO2.
NanoBRET Nano-Glo substrate (Promega) was added to both control and experimental samples at a final concentration of 10 µM. Readings were performed within 10 minutes using a ClarioSTAR (BMG labtech) equipped with 460 nm and 610 nm filters. A corrected BRET ratio was calculated and is defined as the ratio of the emission at 610 nm/460 nm for experimental samples minus the emission at 610 nm/460 nm for control samples (without NanoBRET fluorescent ligand). BRET ratios are expressed as milliBRET units (mBU), where 1 mBU corresponds to the corrected BRET ratio multiplied by 1000.
The co-crystal structure of Spin1 with VinSpinIn confirms that VinSpinIn binds to both domain 1 and 2 of Spin1, and therefore is referred to as a bidentate inhibitor (Figure 1).
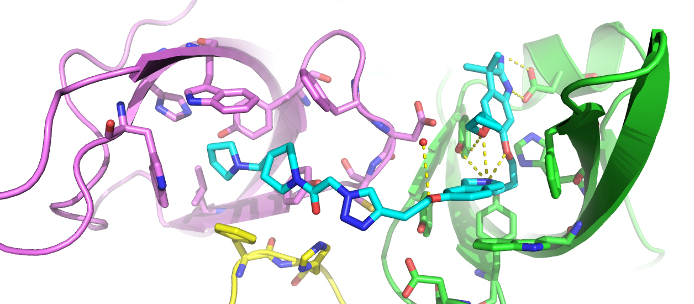
Figure 1: Co-crystal structure of Spin1 (domain 1 = purple; domain 2 = green; domain 3 = yellow) with VinSpinIn (Cyan).