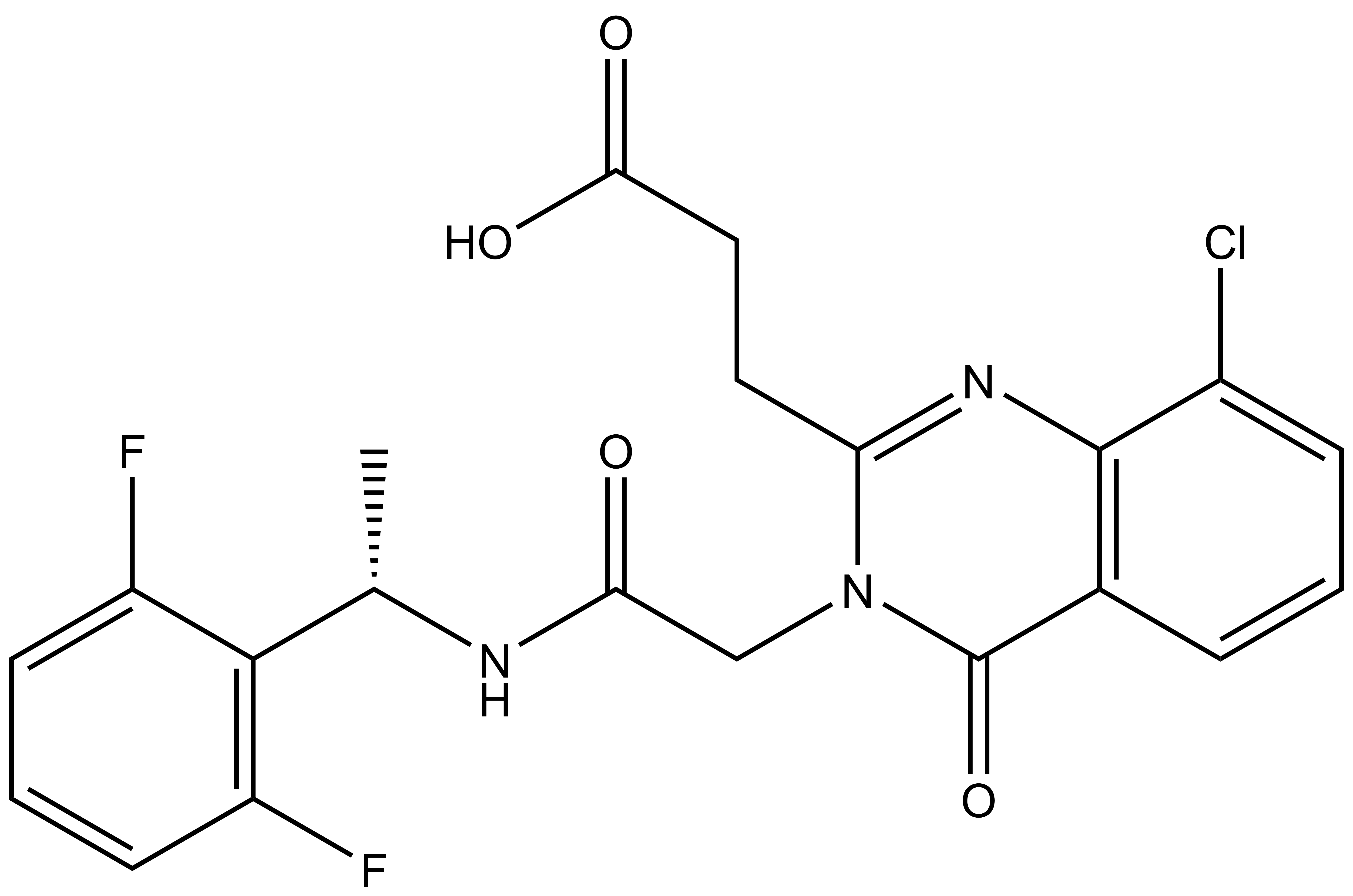
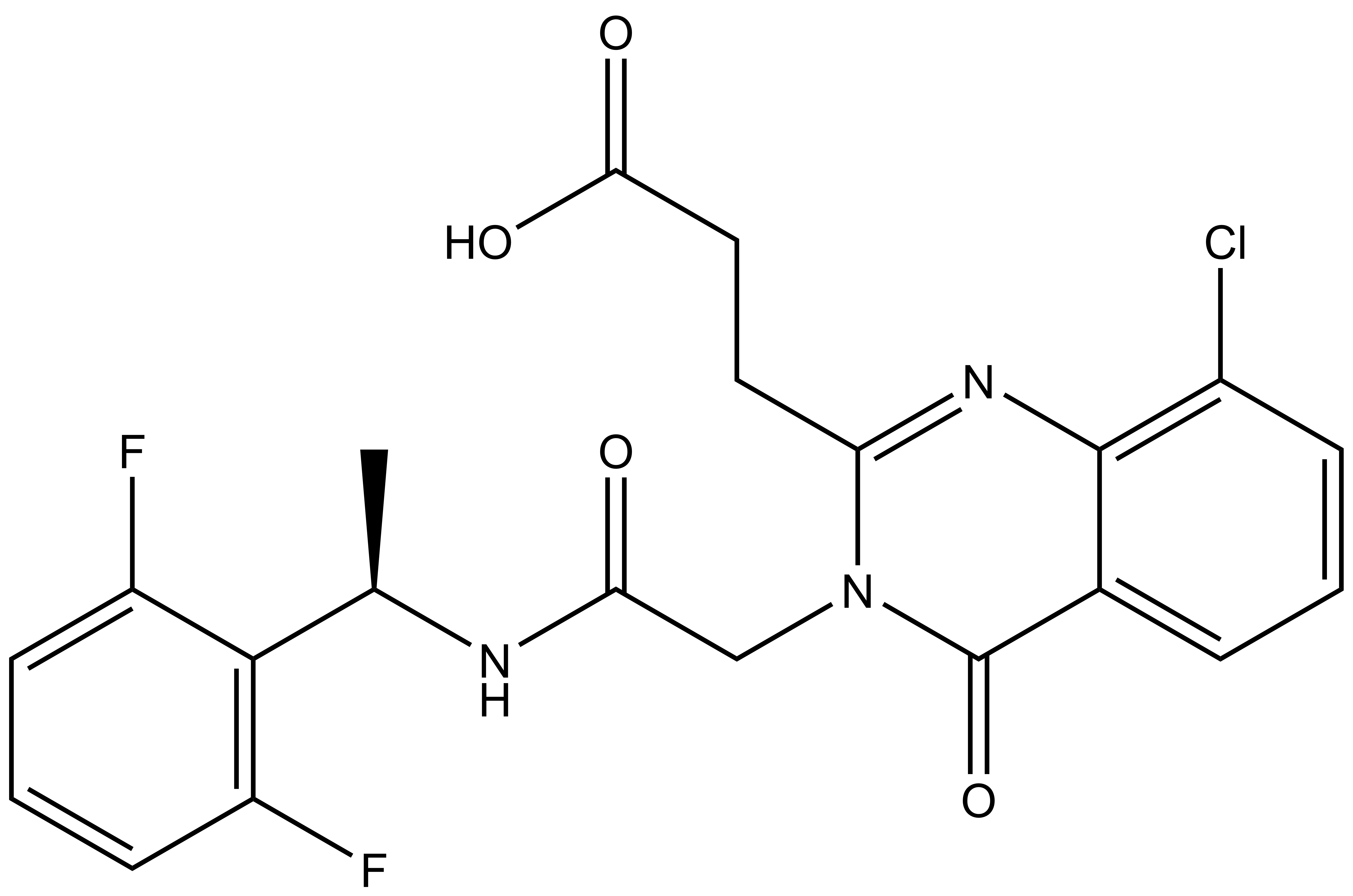
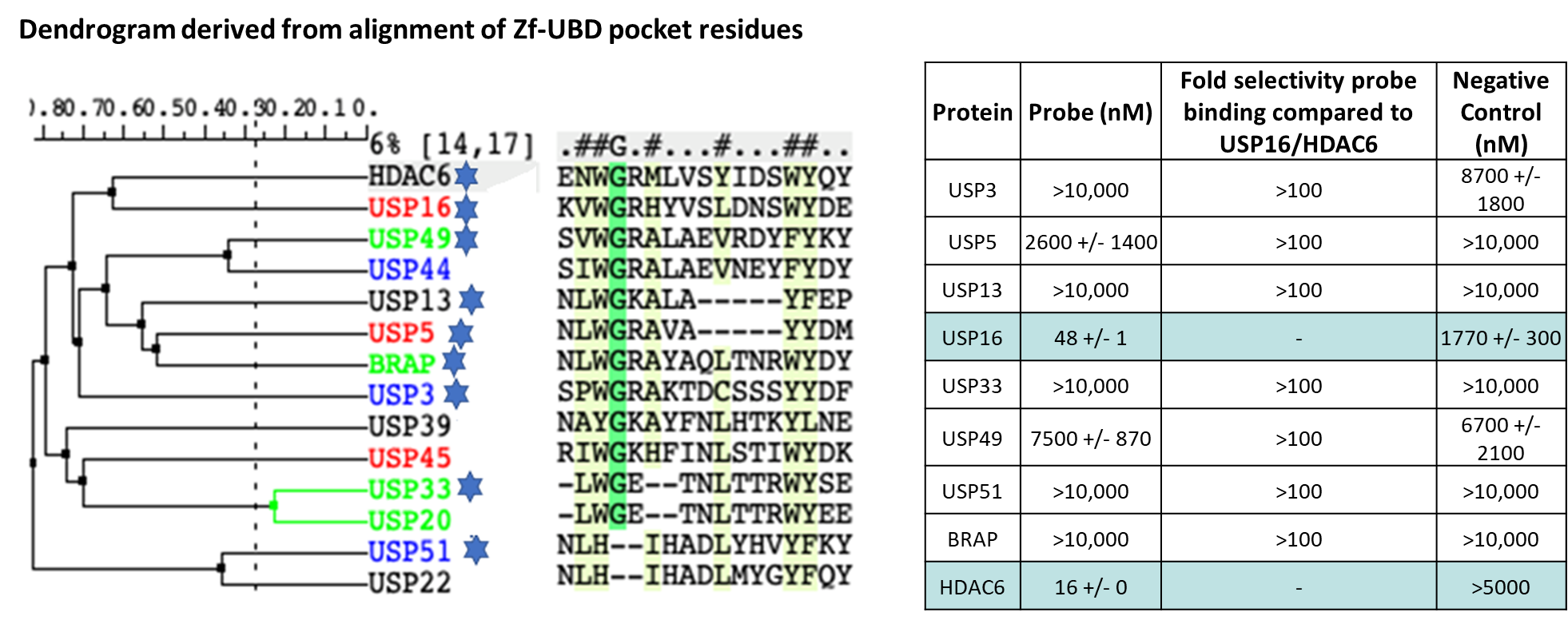
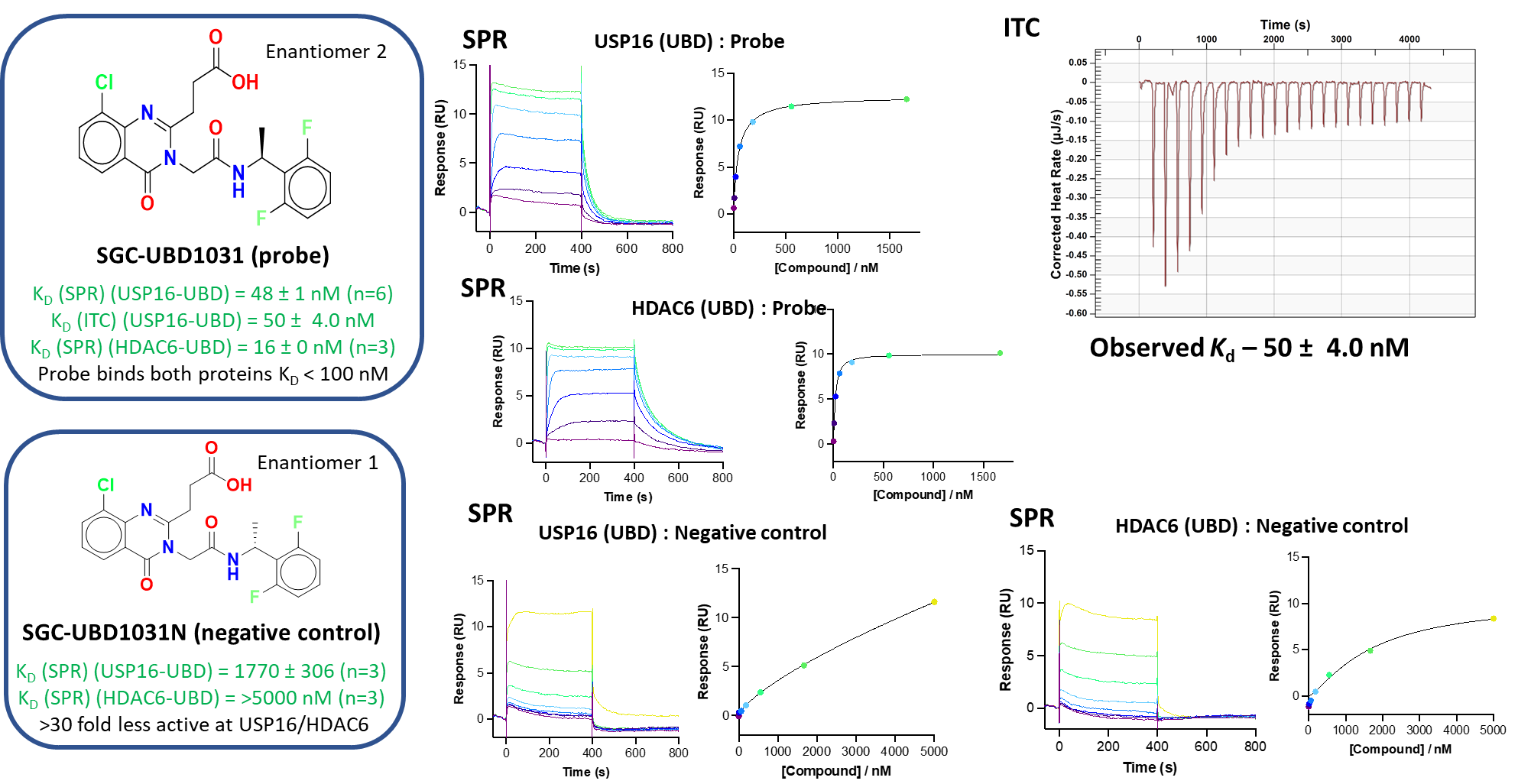
Shaping Tomorrow: The Women Leading Open Science Advancements at the Structural Genomics Consortium [Part 2]
Imagine being part of a world that shapes the future yet feeling invisible within it. This is the reality for women in STEM, who are nearly twice as likely as those in other fields to contemplate leaving their careers behind. The culprits? Overwhelming burnout, overlooked achievements, wage disparities, and a yearning for work that resonates on a deeper level.
To celebrate the International Day of Women and Girls in STEM earlier this year, I had the chance to interview a dynamic and global network of female scientists, who actively work to accelerate early drug discovery.