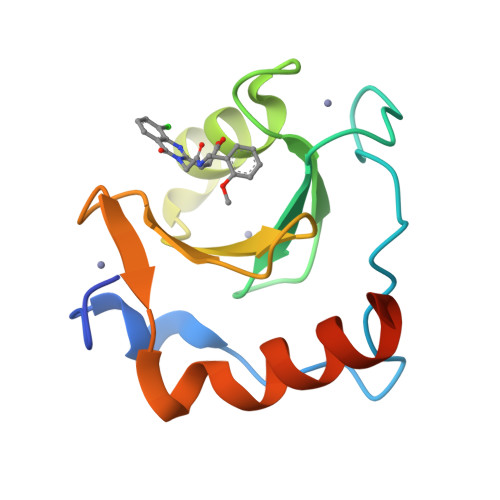
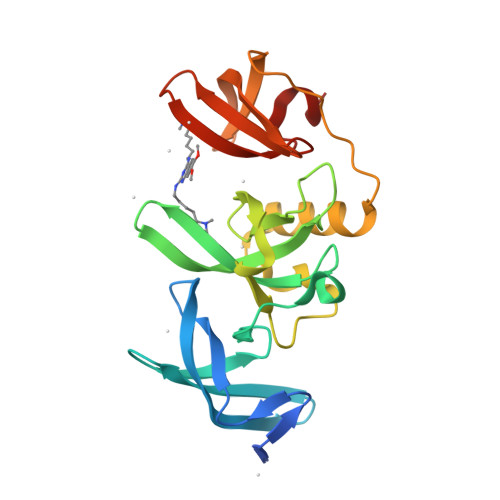
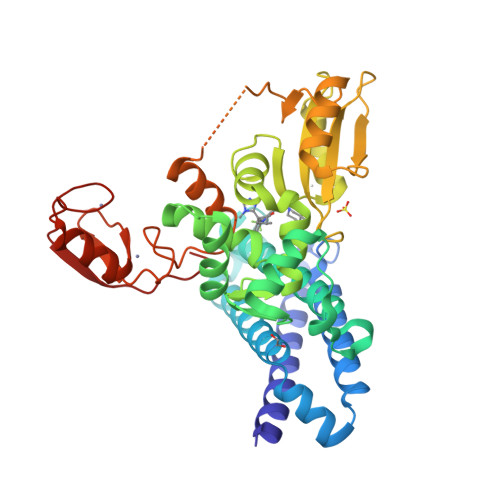
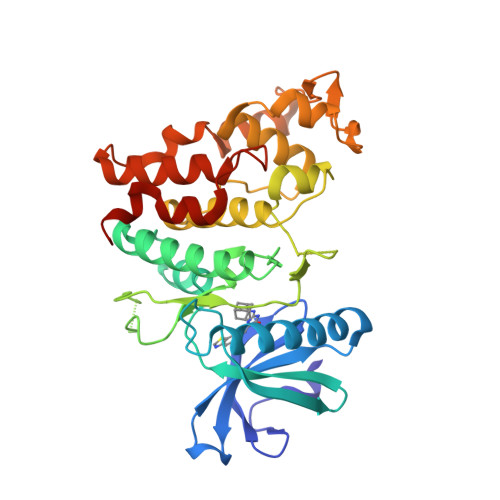
Protein:
HDAC6
PDB ID:
8G43
Deposition Date:
Wednesday 8th February 2023
Authors:
Harding, R.J., Franzoni, I., Mann, M.K., Szewczyk, M., Mirabi, B., Owens, D.D.G., Ackloo, S., Scheremetjew, A., Juarez-Ornelas, K.A., Sanichar, R., Baker, R.J., Dank, C., Brown, P.J., Barsyte-Lovejoy, D., Santhakumar, V., Schapira, M., Lautens, M., Arrowsmith, C.H.