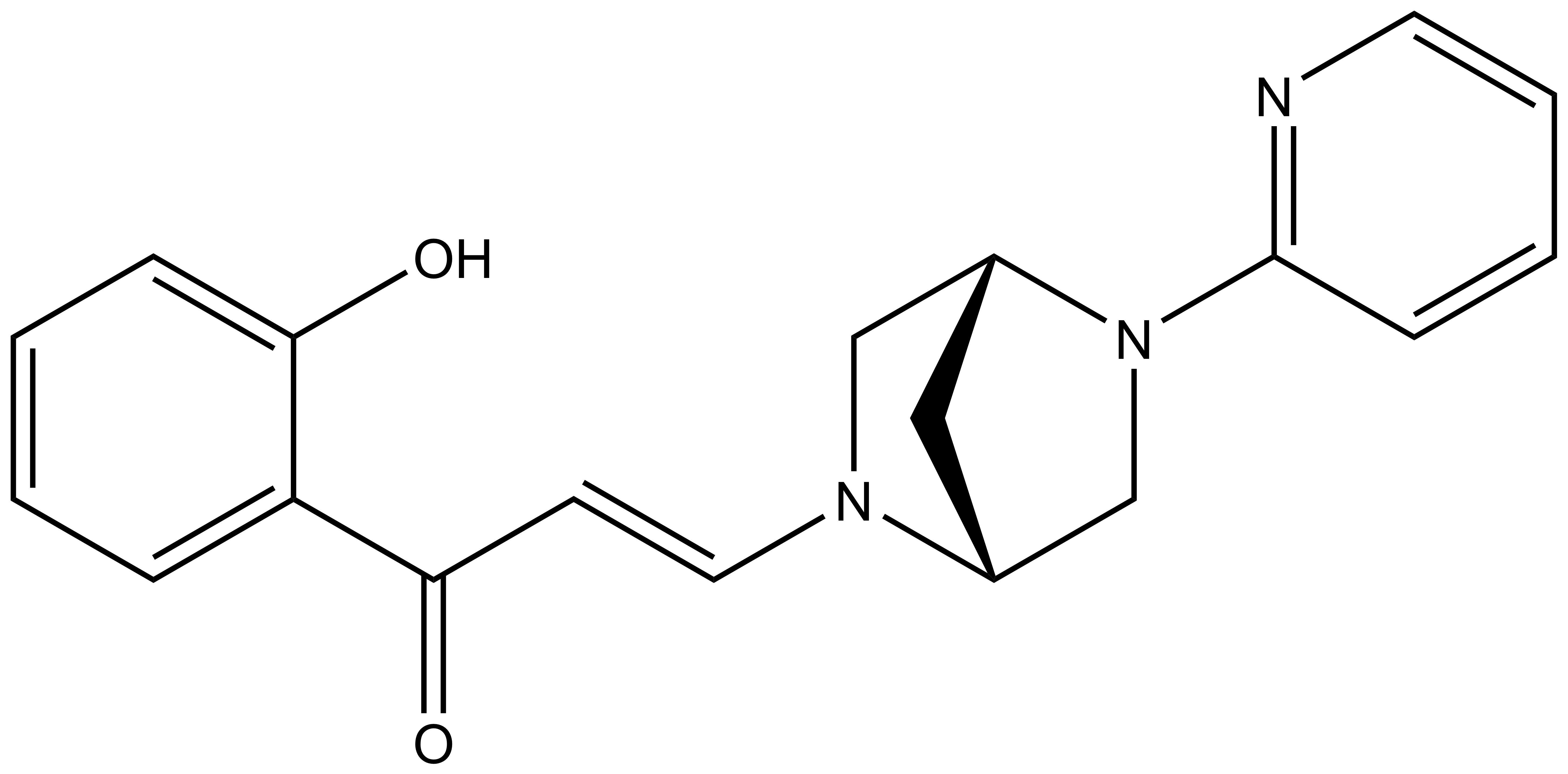
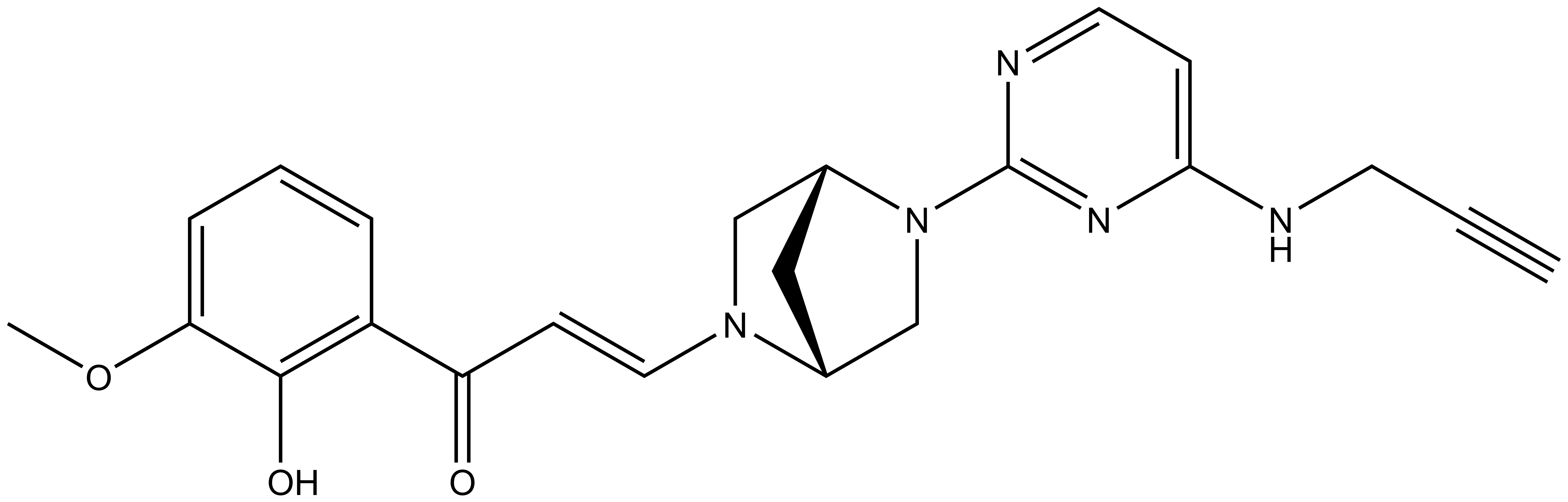
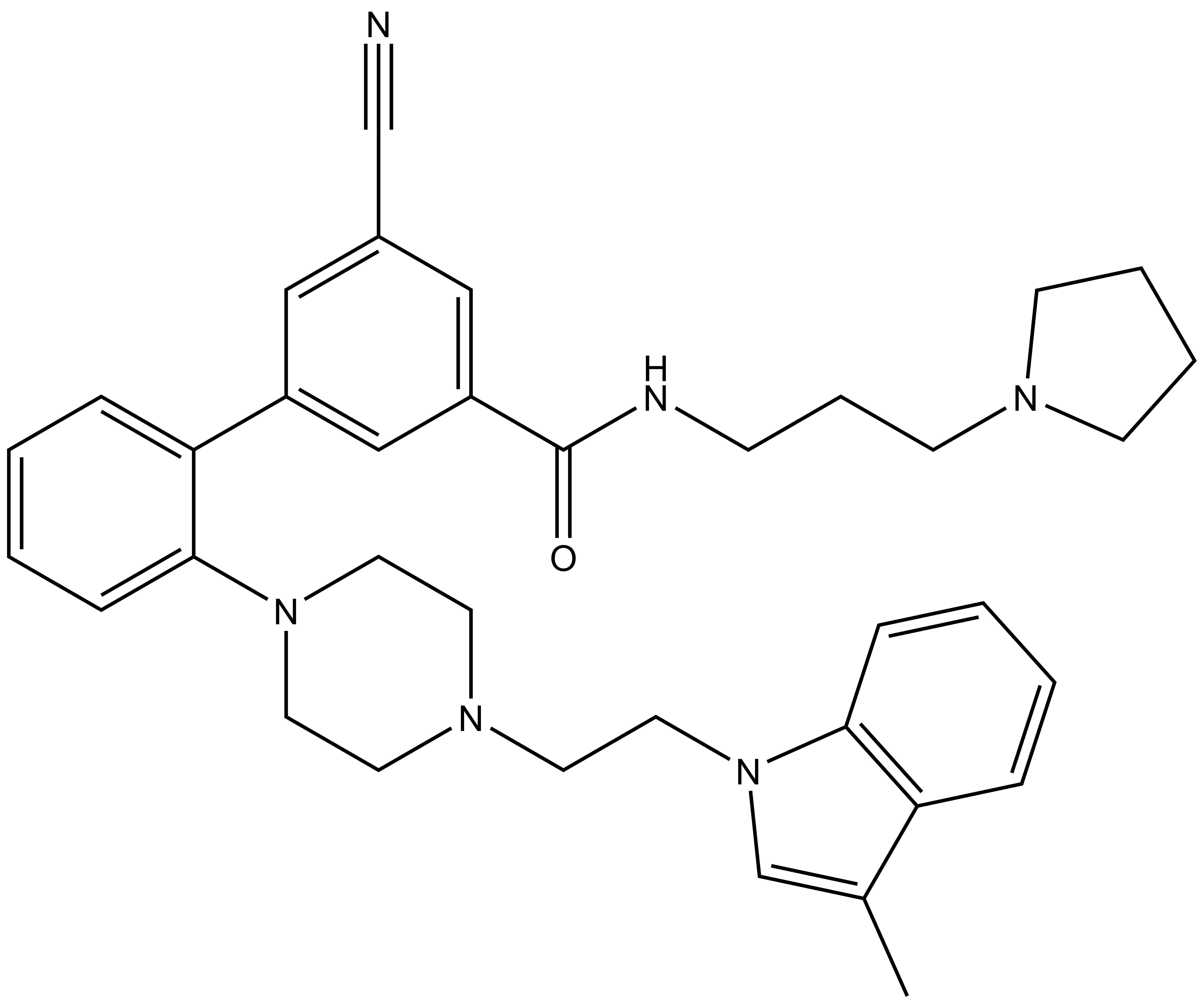
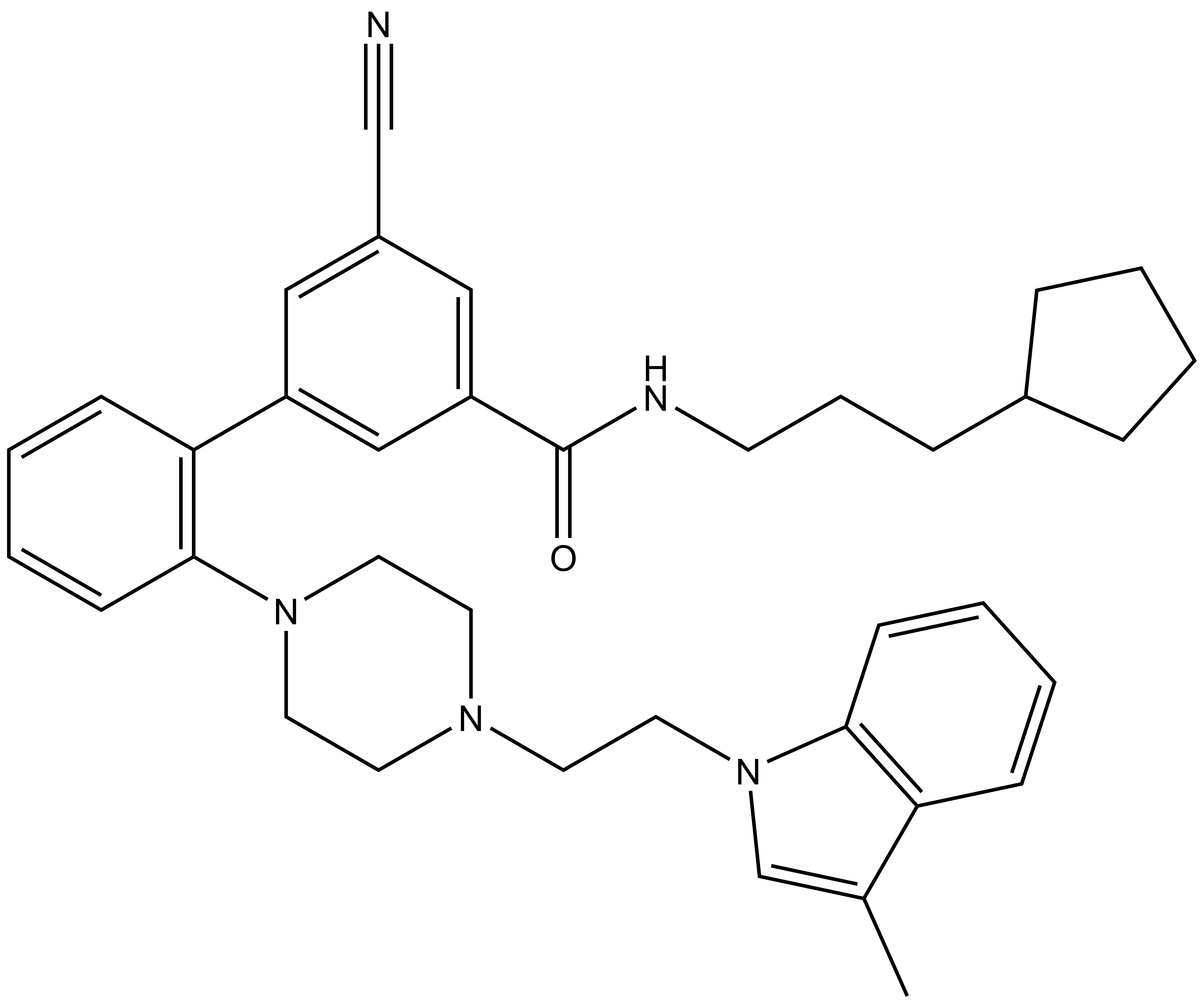
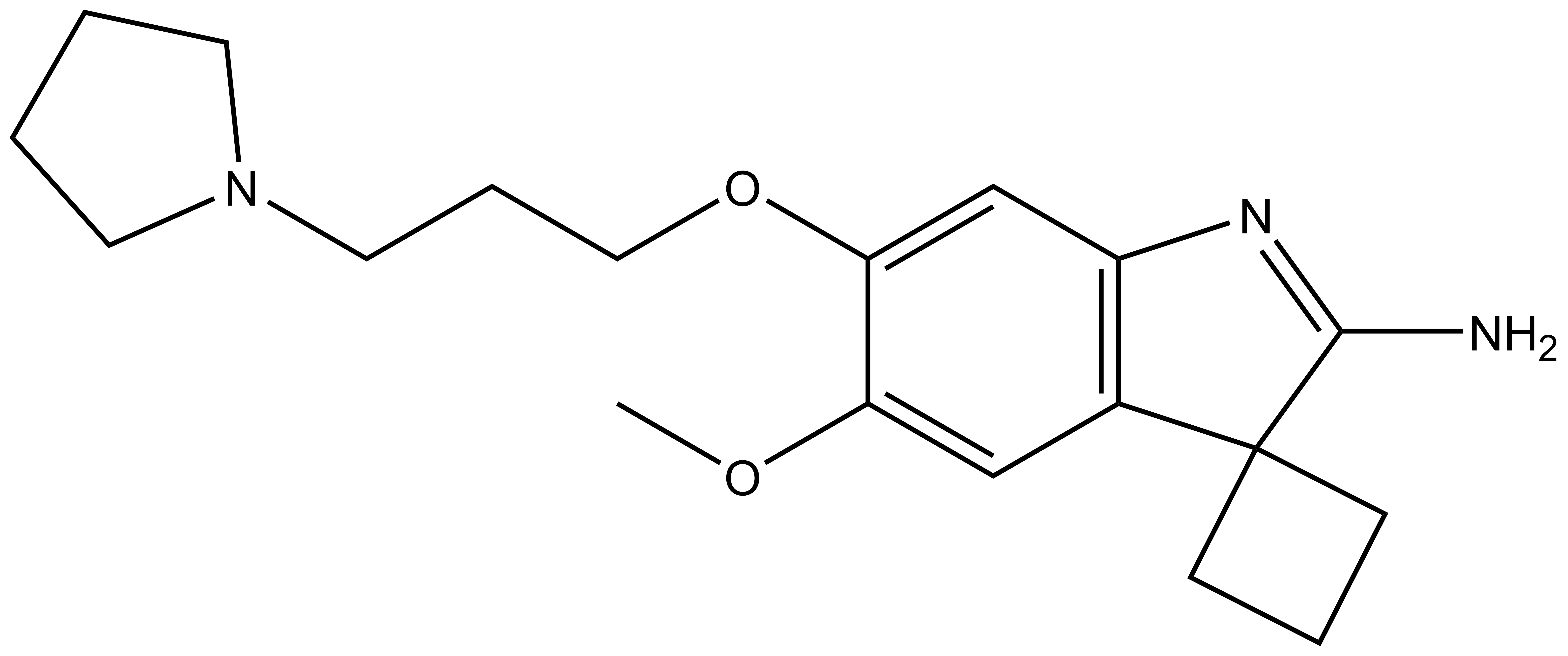
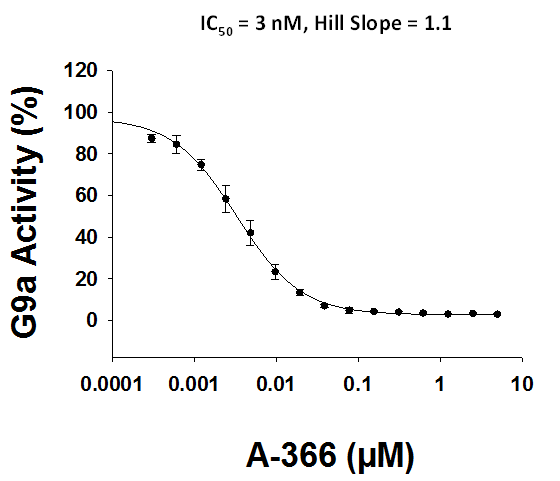
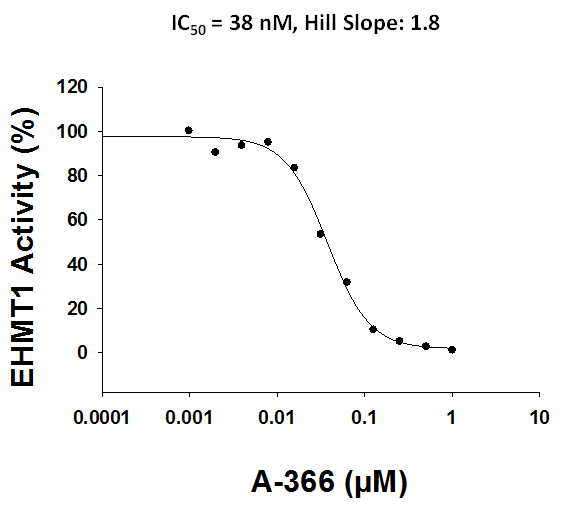
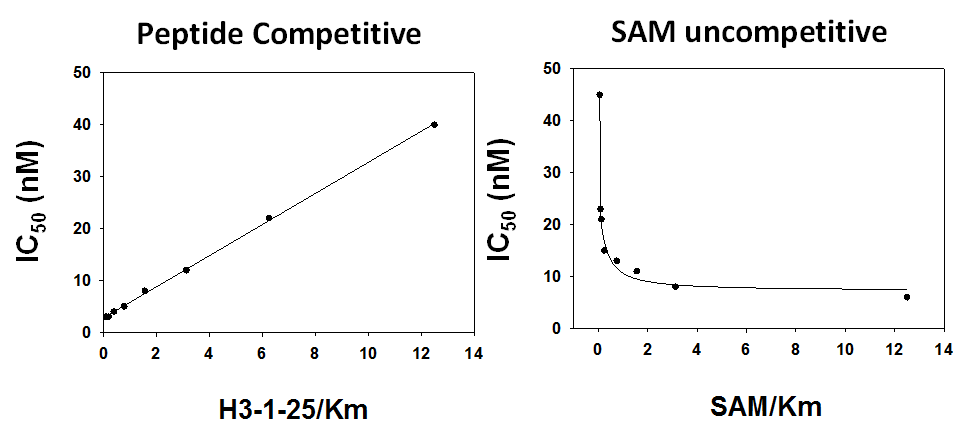
Bayer HealthCare to Join Not-For-Profit SGC to Accelerate Research in Epigenetics
Toronto, December 18, 2013 – Bayer Inc. is pleased to announce that Bayer HealthCare has joined the Structural Genomics Consortium (SGC), a not-for-profit, public-private partnership with active research facilities at the Universities of Toronto and Oxford, UK. Bayer will support funding the consortium to accelerate precompetitive drug research in the areas of protein sciences and epigenetics. Furthermore, Bayer will provide a subset of its compound library for screening to the SGC and will also conduct the chemical work to identify probes.