| Probe | | Negative control |
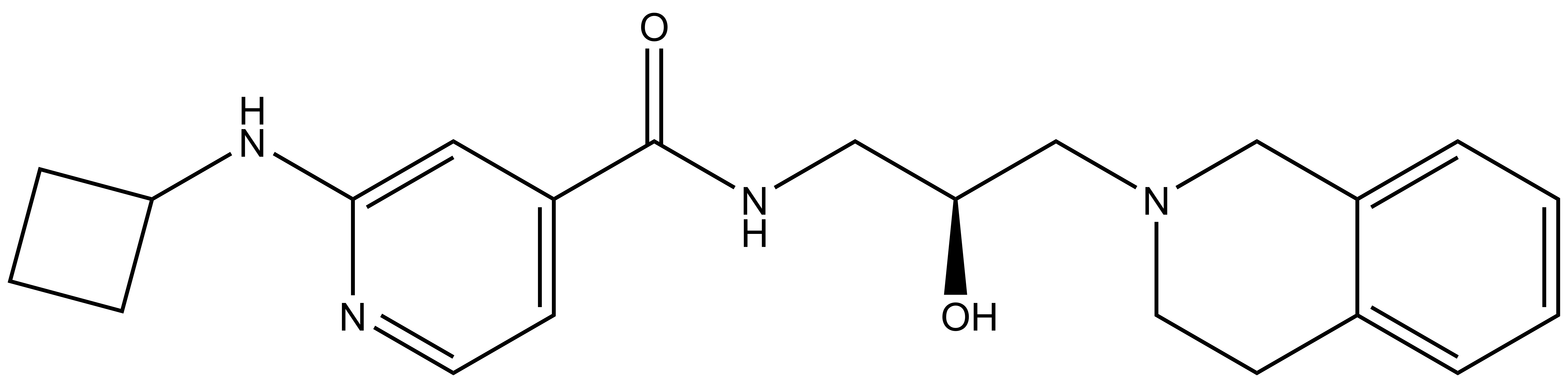 | | 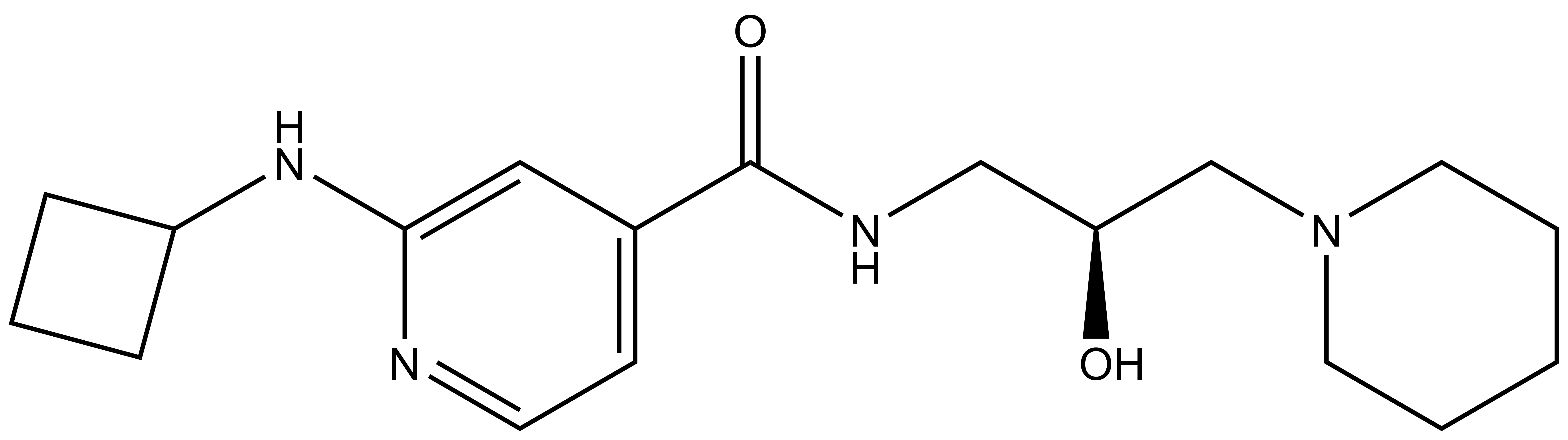 |
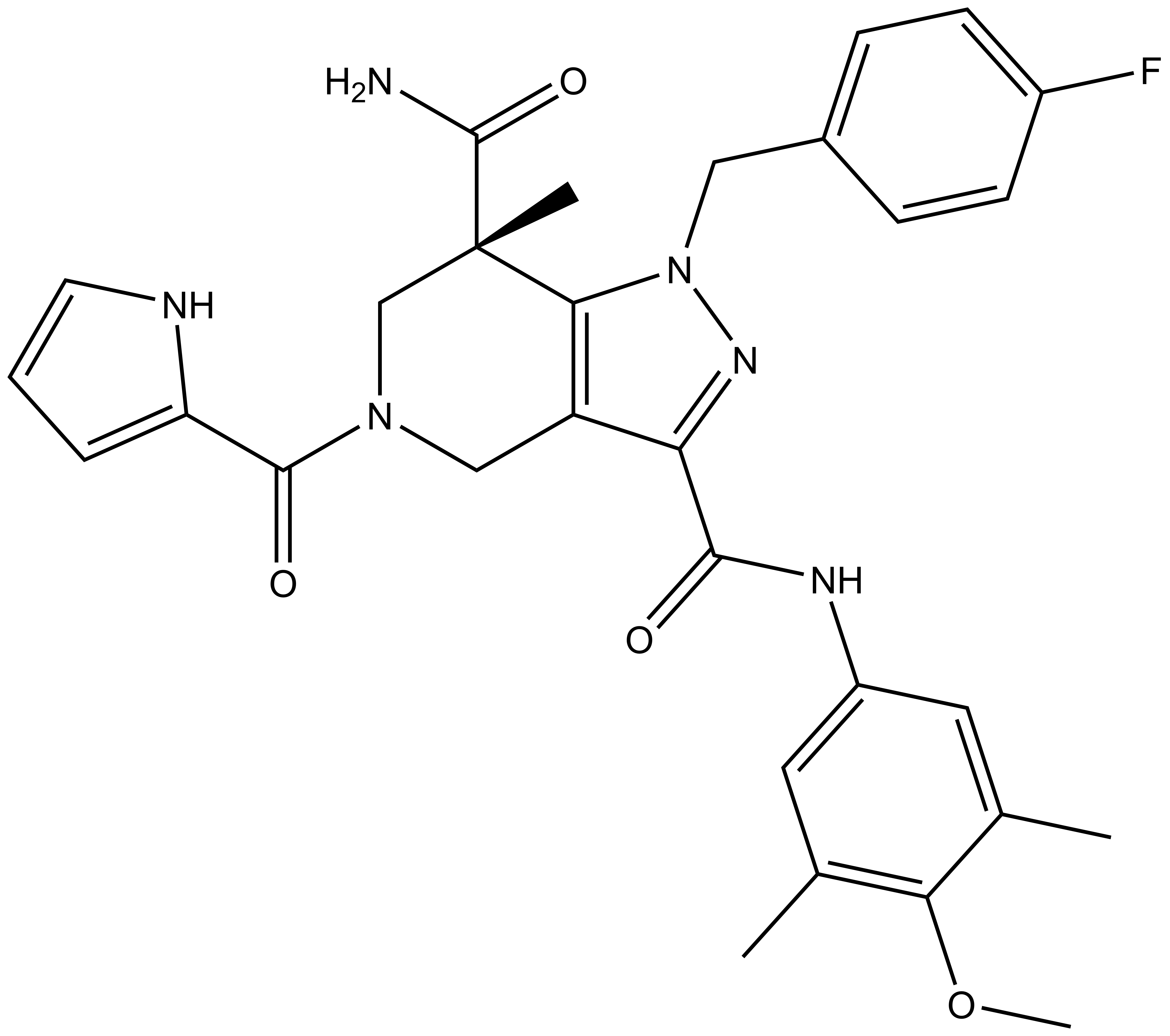
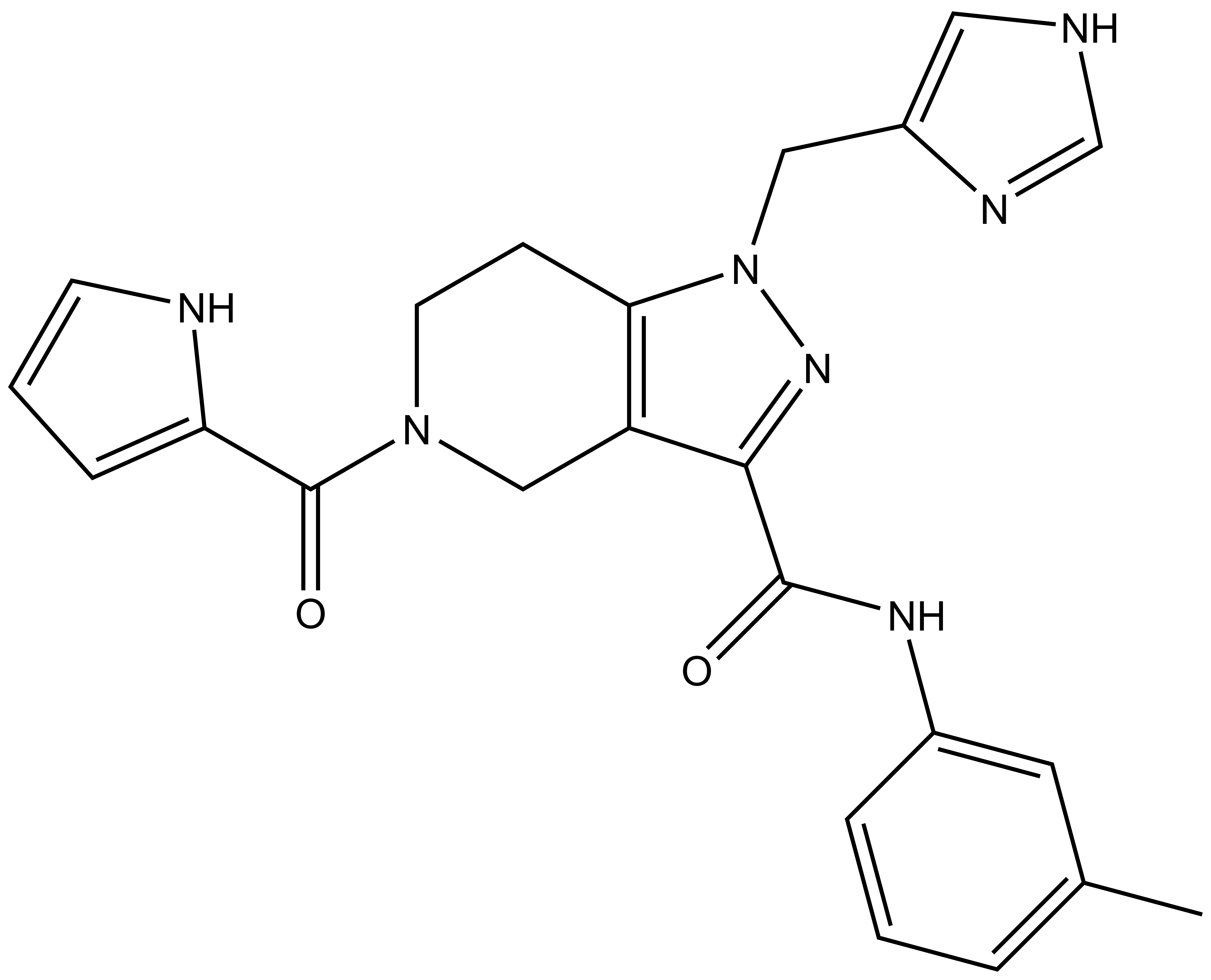
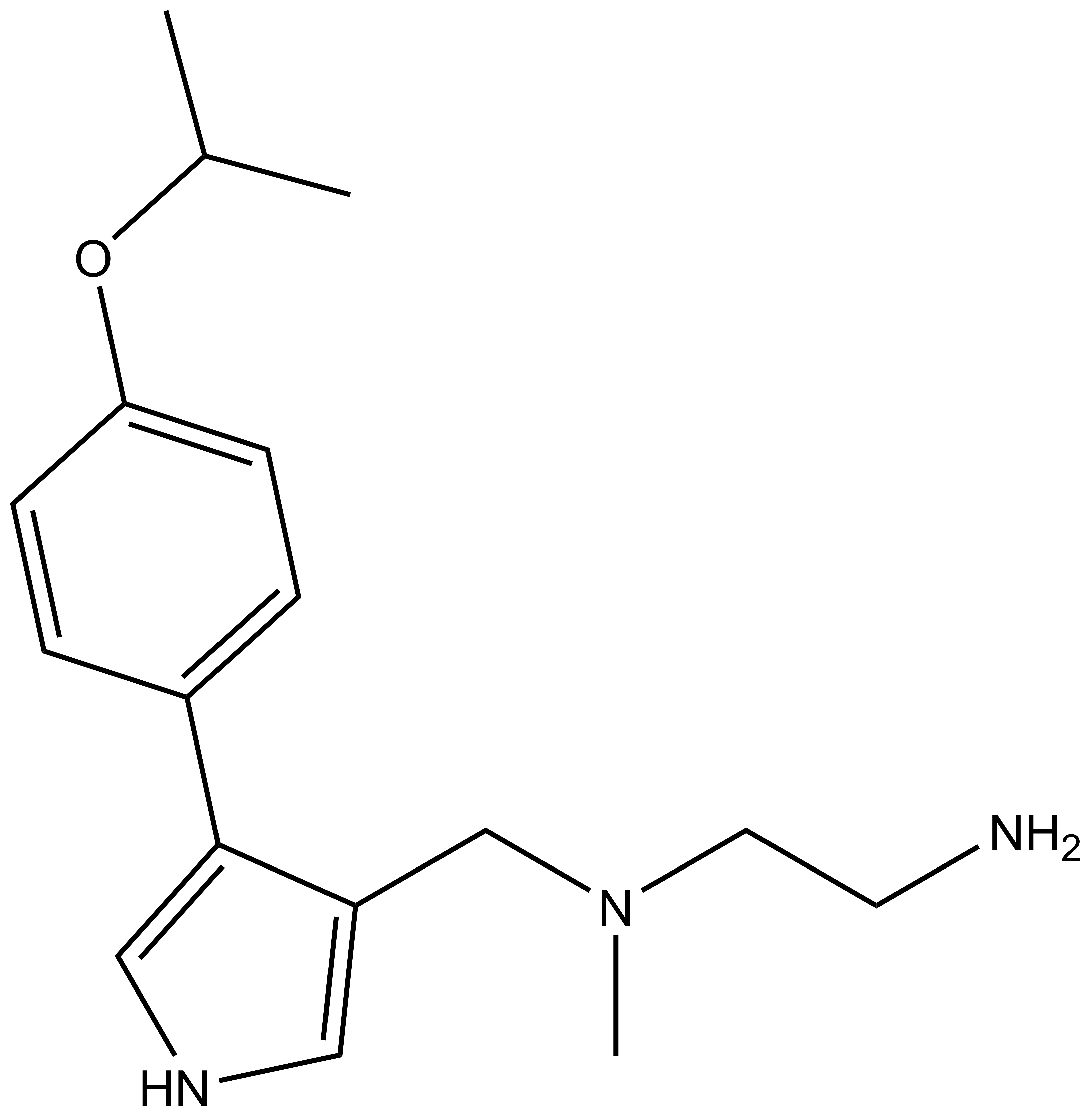
GSK591 | | SGC2096 |
| Physical and chemical properties for GSK591 |
| Molecular weight | 380.2 |
| Molecular formula | C22H28N4O2 |
| IUPAC name | (3-(3-aza-bicyclo[4.4.0]deca-1(10),6,8-trien-3-yl)-2-hydroxy-propylamino)-(2-cyclobutylamino-pyridin-4-yl)-methanone |
| MollogP | 2.311 |
| PSA | 64.84 |
| No. of chiral centres | 1 |
| No. of rotatable bonds | 8 |
| No. of hydrogen bond acceptors | 5 |
| No. of hydrogen bond donors | 3 |
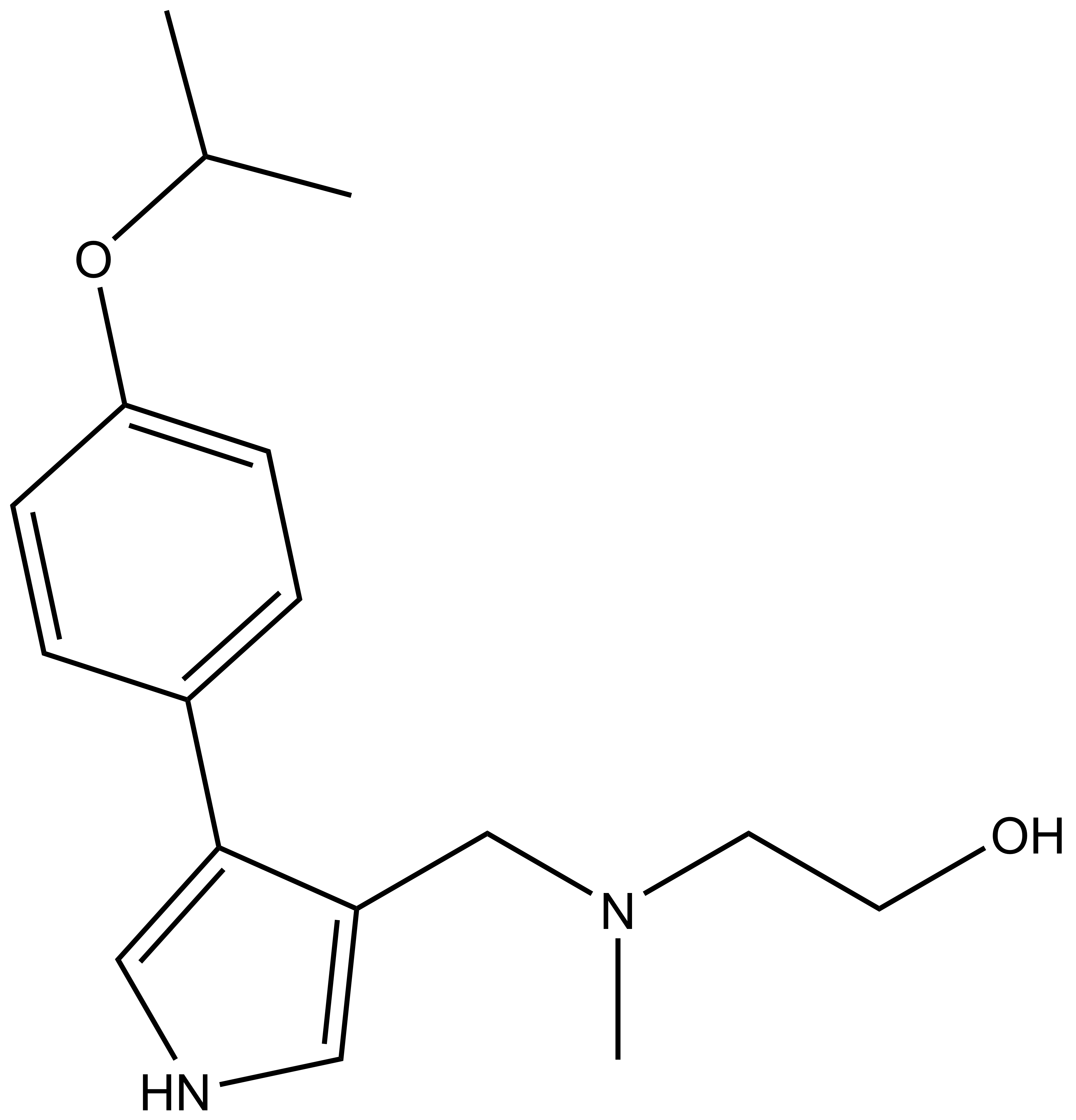
| Physical and chemical properties for SGC2096 |
| Molecular weight | 332.2 |
| Molecular formula | C18H28N4O2 |
| IUPAC name | (2-cyclobutylamino-pyridin-4-yl)-(2-hydroxy-3-(piperidin-1-yl)-propylamino)-methanone |
| MollogP | 1.991 |
| PSA | 65.18 |
| No. of chiral centres | 1 |
| No. of rotatable bonds | 8 |
| No. of hydrogen bond acceptors | 5 |
| No. of hydrogen bond donors | 3 |
- SMILES:
- GSK591: O[C@H](CN1CCC2=CC=CC=C2C1)CNC(C3=CC=NC(NC4CCC4)=C3)=O
- SGC2096: O=C(C1=CC=NC(NC2CCC2)=C1)NC[C@H](O)CN3CCCCC3
- InChI:
- GSK591: InChI=1S/C22H28N4O2/c27-20(15-26-11-9-16-4-1-2-5-18(16)14-26)13-24-22(28)17-8-10-23-21(12-17)25-19-6-3-7-19/h1-2,4-5,8,10,12,19-20,27H,3,6-7,9,11,13-15H2,(H,23,25)(H,24,28)/t20-/m0/s1
- SGC2096: InChI=1S/C18H28N4O2/c23-16(13-22-9-2-1-3-10-22)12-20-18(24)14-7-8-19-17(11-14)21-15-5-4-6-15/h7-8,11,15-16,23H,1-6,9-10,12-13H2,(H,19,21)(H,20,24)/t16-/m0/s1
- InChIKey:
- GSK591: TWKYXZSXXXKKJU-FQEVSTJZSA-N
- SGC2096: DCHQEWPVLGTADW-INIZCTEOSA-N
| Biotinylated inhibitor |
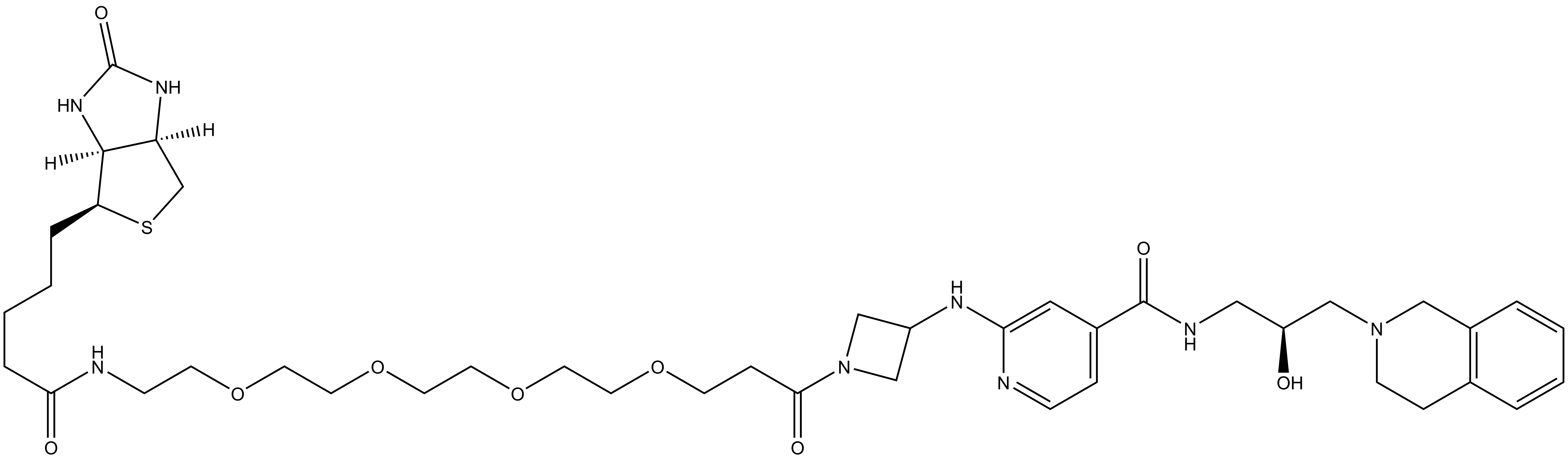 |
SGC3185 |
| Physical and chemical properties for GSK3185 |
| Molecular weight | 854.4 |
| Molecular formula | C42H62N8O9S |
| IUPAC name | 6-(5-(2-(2-(2-(2-(3-(3-(4-((3-(3-aza-bicyclo[4.4.0]deca-1(10),6,8-trien-3-yl)-2-hydroxy-propylamino)-formyl)-pyridin-2-ylamino)-azetidin-1-yl)-3-oxo-propoxy)-ethoxy)-ethoxy)-ethoxy)-ethylamino)-5-oxo-pentyl)-7-thia-2,4-diaza-bicyclo[3.3.0]octan-3-one |
| MollogP | 1.341 |
| PSA | 175.1 |
| No. of chiral centres | 4 |
| No. of rotatable bonds | 30 |
| No. of hydrogen bond acceptors | 16 |
| No. of hydrogen bond donors | 6 |
SMILES: [H][C@@]12[C@H](CCCCC(NCCOCCOCCOCCOCCC(N3CC(C3)Nc3cc(ccn3)C(NC[C@@H](CN3CCc4ccccc4C3)O)=O)=O)=O)SC[C@]1([H])NC(N2)=O
InChI: InChI=1S/C42H62N8O9S/c51-34(28-49-14-10-30-5-1-2-6-32(30)25-49)24-45-41(54)31-9-12-43-37(23-31)46-33-26-50(27-33)39(53)11-15-56-17-19-58-21-22-59-20-18-57-16-13-44-38(52)8-4-3-7-36-40-35(29-60-36)47-42(55)48-40/h1-2,5-6,9,12,23,33-36,40,51H,3-4,7-8,10-11,13-22,24-29H2,(H,43,46)(H,44,52)(H,45,54)(H2,47,48,55)/t34-,35-,36-,40-/m0/s1
InChIKey: PZIWNLNYZRZJCY-VNLYVSNMSA-N