
Dr. Dalia Barsyte-Lovejoy, PhD is an Assistant Professor at the Department of Pharmacology and Toxicology, UofT, and Principal Investigator at the SGC-Toronto, working to understand fundamental regulatory mechanisms of epigenetic proteins and their pharmacological modulation in cancer. The group’s research focuses on disease mechanisms, therapeutic targets, and chemical probe discovery, resulting in over 30 extensively characterized compounds that have helped shape the emerging field of epigenetics and enabled over 50 collaborative projects that are uncovering new epigenetic mechanisms in cancer and its treatment.
We are interested in understanding the mechanism of epigenetic regulators and posttranslational modifications that control cancer cell growth, differentiation, and therapy response. Protein lysine and arginine methyltransferases regulate transcription, genome stability, splicing, RNA metabolism, and other cell processes dictated by which substrates these enzymes methylate. Lysine methyltransferases such as EZH2 and NSD2 primarily methylate histones to establish repressive and active chromatin. In contrast, arginine methyltransferases have a broad scope of substrates ranging from histones to signaling molecules, enzymes, and structural proteins. Epigenetic chromatin regulation, transcriptome, and cellular signaling are fine-tuned by ubiquitin modification. Our work seeks to understand how these posttranslational modifications are misregulated in cancer and identify new therapeutic targets.
Through multidisciplinary research that includes cell and chemical biology, protein structural biology, and many collaborative studies with colleagues across industry and academia, the SGC chemical probes project has generated several probes for methyltransferases, ubiquitin ligases, and deubiquitylases. We are currently using these chemical probes to explore the cellular pathways in poor prognosis acute myeloid leukemia, pancreatic, lung and breast cancer.
Chemical probes as tools for cancer target discovery
|
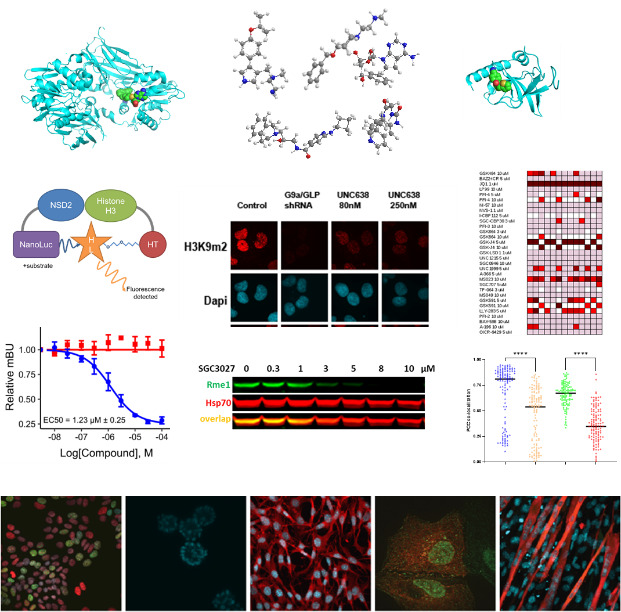
To study epigenetic modifier proteins, we need genetic and pharmacological tools. Chemical probe compounds that potently and selectively inhibit or degrade the target proteins in cells provide tools for modulating activating/repressing histone marks and other cellular signaling pathways. By discovering and using chemical probes, we expand our understanding of the protein function and its therapeutic utility to establish a biological rationale in cancer therapy.
|
Link to Open Lab notebooks that features science community posts on our various projects https://openlabnotebooks.org/
Shrestha S, Maitland MER, Jing L, Duan S, Nie DY, St-Germain J, Kanaris M, Barsyte-Lovejoy D, Arrowsmith CH, Raught B
Nat Commun. 2025-8-31 . 16(1):8089 .doi: 10.1038/s41467-025-63357-7
PMID: 40883309Kimani SW, Noureldin M, Wilson B, Hoffer L, Green SR, Szewczyk MM, González-Álvarez H, Mohammed M, Chan M, Krausser C, Li ASM, Hajian T, Tucker S, Joshi D, Saraon P, Thériault B, Kim JS, Santhakumar V, Loppnau P, Li Y, Seitova A, Dong A, Kiyota T, Hammann T, Gehrtz P, Patel B, Rathod V, Vala A, Rout B, Jagodra P, Brown PJ, Aman A, Ramnauth J, Poda G, Uehling D, Arrowsmith CH, Barsyte-Lovejoy D, Marcellus R, Ackloo S, Mamai A, Al-Awar R, Halabelian L
Commun Biol. 2025-7-20 . 8(1):1076 .doi: 10.1038/s42003-025-08491-0
PMID: 40683980Aparnathi MK, Ul Haq S, St-Germain J, Nixon KCJ, Walton J, Song L, Majeed S, Patel PS, Subramaniam R, Philip V, Marcellus R, Barsyte-Lovejoy D, Al-Awar R, Hakem R, Arrowsmith CH, Ailles L, Raught B, Lok BH
Neoplasia. 2025-5-26 . 66:101176 .doi: 10.1016/j.neo.2025.101176
PMID: 40413955Yadav M, AlQazzaz MA, Ciamponi FE, Ho JC, Maron MI, Sababi AM, MacLeod G, Ahmadi M, Bullivant G, Tano V, Langley SR, Sánchez-Osuna M, Sachamitr P, Kushida M, Bardile CF, Pouladi MA, Kurtz R, Richards L, Pugh T, Tyers M, Angers S, Dirks PB, Bader GD, Truant R, Massirer KB, Barsyte-Lovejoy D, Shechter D, Harding RJ, Arrowsmith CH, Prinos P
Nucleic Acids Res. 2025-4-30 . 53(8): .doi: 10.1093/nar/gkaf347
PMID: 40304179Uguen M, Shell DJ, Silva M, Deng Y, Li F, Szewczyk MM, Yang K, Zhao Y, Stashko MA, Norris-Drouin JL, Waybright JM, Beldar S, Rectenwald JM, Mordant AL, Webb TS, Herring LE, Arrowsmith CH, Ackloo S, Gygi SP, McGinty RK, Barsyte-Lovejoy D, Liu P, Halabelian L, James LI, Pearce KH, Frye SV
Nat Commun. 2025-2-25 . 16(1):1905 .doi: 10.1038/s41467-025-57005-3
PMID: 39994194Maitland MER, Onea G, Owens DDG, Gonga-Cavé BC, Wang X, Arrowsmith CH, Barsyte-Lovejoy D, Lajoie GA, Schild-Poulter C
Commun Biol. 2024-12-20 . 7(1):1668 .doi: 10.1038/s42003-024-07371-3
PMID: 39702571Mabanglo MF, Wilson B, Noureldin M, Kimani SW, Mamai A, Krausser C, González-Álvarez H, Srivastava S, Mohammed M, Hoffer L, Chan M, Avrumutsoae J, Li ASM, Hajian T, Tucker S, Green S, Szewczyk M, Barsyte-Lovejoy D, Santhakumar V, Ackloo S, Loppnau P, Li Y, Seitova A, Kiyota T, Wang JG, Privé GG, Kuntz DA, Patel B, Rathod V, Vala A, Rout B, Aman A, Poda G, Uehling D, Ramnauth J, Halabelian L, Marcellus R, Al-Awar R, Vedadi M
Nat Commun. 2024-11-24 . 15(1):10165 .doi: 10.1038/s41467-024-54500-x
PMID: 39580491Ackloo S, Li F, Szewczyk M, Seitova A, Loppnau P, Zeng H, Xu J, Ahmad S, Arnautova YA, Baghaie AJ, Beldar S, Bolotokova A, Centrella PA, Chau I, Clark MA, Cuozzo JW, Dehghani-Tafti S, Disch JS, Dong A, Dumas A, Feng JA, Ghiabi P, Gibson E, Gilmer J, Goldman B, Green SR, Guié MA, Guilinger JP, Harms N, Herasymenko O, Houliston S, Hutchinson A, Kearnes S, Keefe AD, Kimani SW, Kramer T, Kutera M, Kwak HA, Lento C, Li Y, Liu J, Loup J, Machado RAC, Mulhern CJ, Perveen S, Righetto GL, Riley P, Shrestha S, Sigel EA, Silva M, Sintchak MD, Slakman BL, Taylor RD, Thompson J, Torng W, Underkoffler C, von Rechenberg M, Walsh RT, Watson I, Wilson DJ, Wolf E, Yadav M, Yazdi AK, Zhang J, Zhang Y, Santhakumar V, Edwards AM, Barsyte-Lovejoy D, Schapira M, Brown PJ, Halabelian L, Arrowsmith CH
J Med Chem. 2024-11-4 . .doi: 10.1021/acs.jmedchem.4c02010
PMID: 39495097Sellars E, Savguira M, Wu J, Cancelliere S, Jen M, Krishnan R, Hakem A, Barsyte-Lovejoy D, Hakem R, Narod SA, Kotsopoulos J, Salmena L
iScience. 2024-7-19 . 27(7):110180 .doi: 10.1016/j.isci.2024.110180
PMID: 38993666Nie DY, Tabor JR, Li J, Kutera M, St-Germain J, Hanley RP, Wolf E, Paulakonis E, Kenney TMG, Duan S, Shrestha S, Owens DDG, Maitland MER, Pon A, Szewczyk M, Lamberto AJ, Menes M, Li F, Penn LZ, Barsyte-Lovejoy D, Brown NG, Barsotti AM, Stamford AW, Collins JL, Wilson DJ, Raught B, Licht JD, James LI, Arrowsmith CH
Nat Chem Biol. 2024-7-4 . .doi: 10.1038/s41589-024-01660-y
PMID: 38965384