This probe (hydrochloride) is available from Cayman Chemical, Sigma and Tocris.
| Probe | Negative control | |
 |
|  |
TP-238 |
| TP-422 |
CECR2 (cat eye syndrome chromosome region, candidate 2) gene is predominantly expressed in the nervous system and involved in neurulation. It is located in the segment of chromosome 22q11.2. Multiplication of this segment lead to rare genetic disorder called cat eye syndrome characterized by multiple congenital defects (1). CECR2 has also been implicated in regulation of DNA damage response (2)
Bromodomain PHD finger transcription factor BPTF/FALZ is a core component of the conserved, multi-subunit nucleosome remodelling factor (NURF) complex. BPTF is essential for neural development and haematopoiesis (3). BPTF is involved in c-MYC chromatin recruitment and transcriptional activity in hematopetic and also cancer stem cells (4).
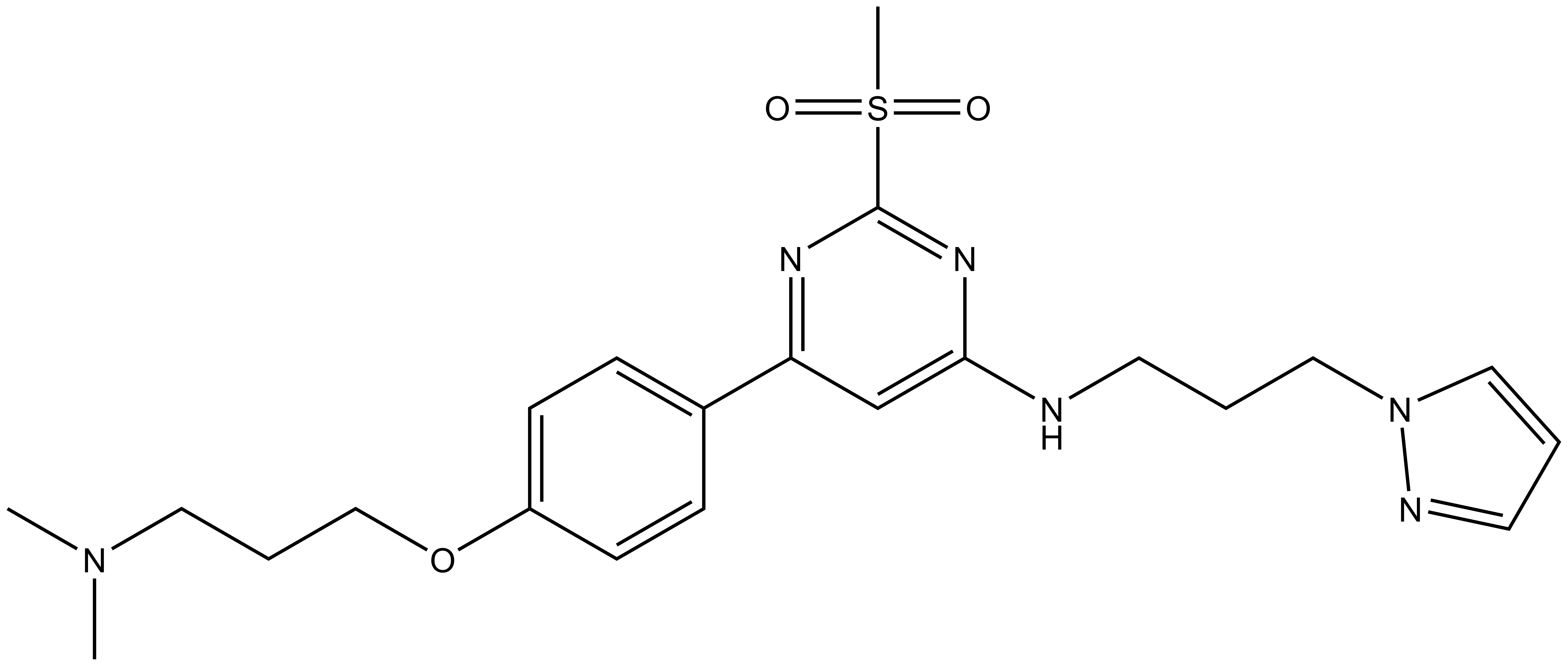
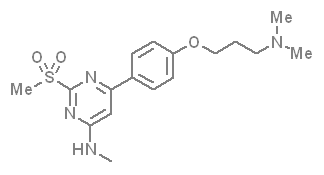
In a collaborative effort Takeda and the SGC have identified and characterised TP-238 as a CECR2/BPTF chemical probe.
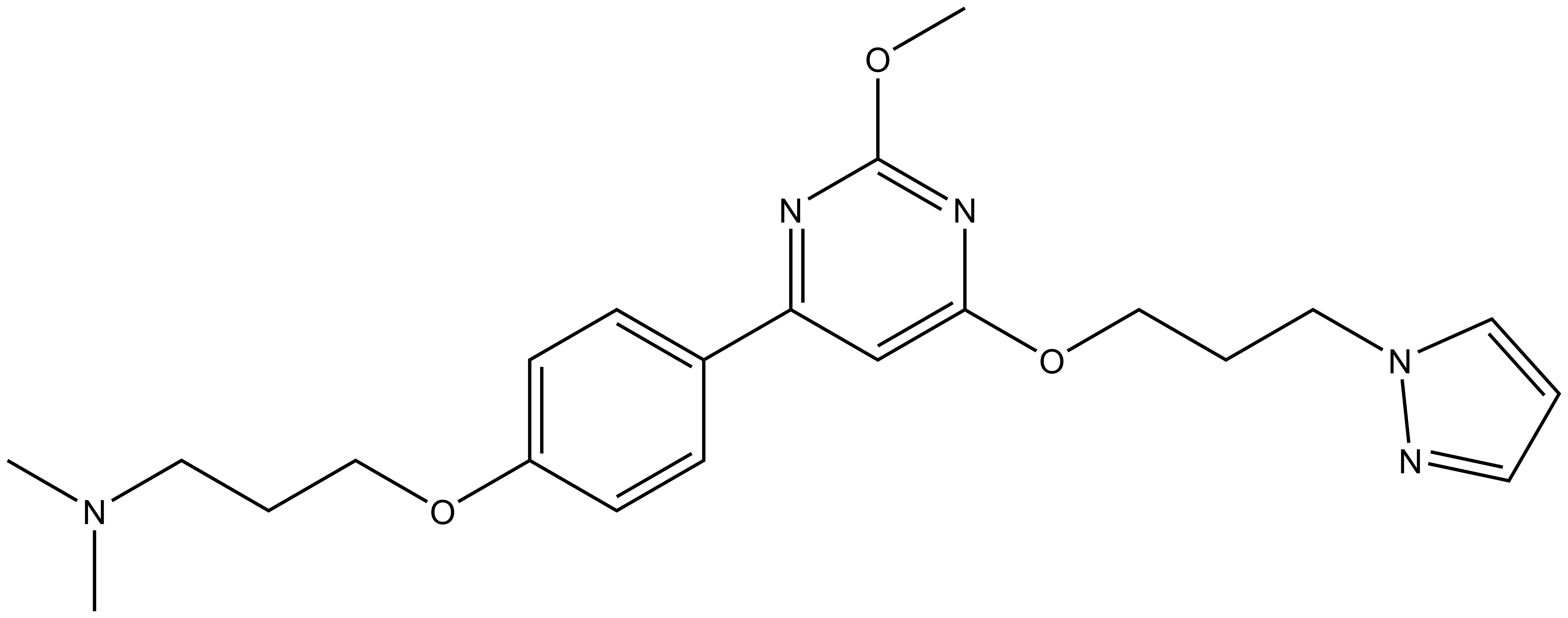
TP-238 has on target biochemical activity of 10-30 nM with CECR2 and 100-350 nM with BPTF. Negative control TP-422 is completely inactive against BPTF and CECR2.
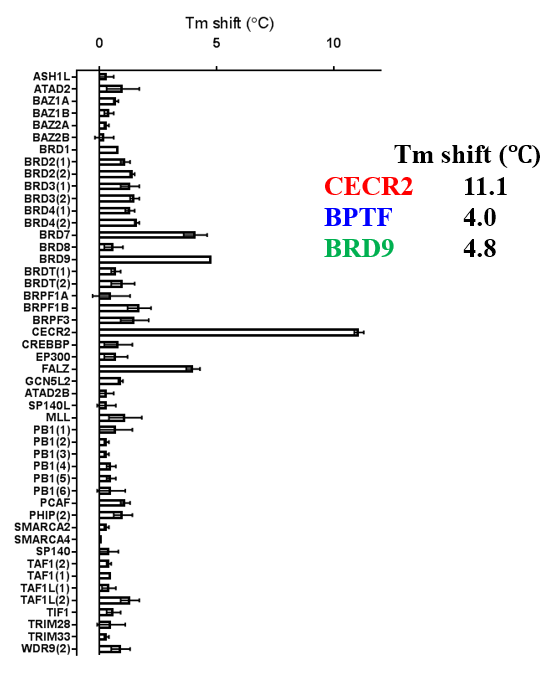
The closest off-target bromodomain inhibition is BRD9 with IC50 of 1.4 µM. TP-238 has been profiled against the panel of 338 kinases and showed no activity at 1 μM.
We recommend that TP-238 and TP-422 be used at no more than 2 µM concentration in cells.
Cell-based NanoBRETTM experiments measured the target engagement with both BPTF and CECR2 with EC50 in the 200-300 nM range.
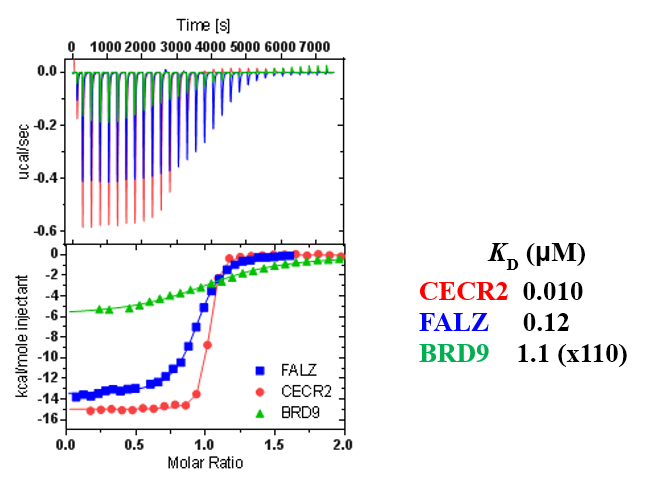
TP-238 shows an IC50 of 30nM against CECR2 and 350nM against BPTF in an alphascreen assay. ITC shows TP-238 with a KD of 10nM for CECR2 and 120nM for BPTF.
| TP-238 |
 |
Click here to download the SDF file. |
SMILES:
CN(CCCOC1=CC=C(C2=NC(S(C)(=O)=O)=NC(NCCCN3C=CC=N3)=C2)C=C1)C
InChI:
InChI=1S/C22H30N6O3S/c1-27(2)13-6-16-31-19-9-7-18(8-10-19)20-17-21(26-22(25-20)32(3,29)30)23-11-4-14-28-15-5-12-24-28/h5,7-10,12,15,17H,4,6,11,13-14,16H2,1-3H3,(H,23,25,26)
InChIKey:
MSIJJXOWLFOYIN-UHFFFAOYSA-N
| Physical and chemical properties | |
| Molecular weight | 458.21 |
| Molecular formula | C22 H30 N6 O3 S |
| IUPAC name | 1-(3-(6-(4-(3-dimethylamino-propoxy)-phenyl)-2-methylsulfonyl-pyrimidin-4-ylamino)-propyl)-1H-pyrazole |
| clogP | 2.4 |
| PSA | 84.5 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 12 |
| No. of hydrogen bond acceptors | 9 |
| No. of hydrogen bond donors | 1 |
| PAMPA (nm/sec, pH=7.4) | 28 |
| Aqueous solubility (µM, pH= 6.8) | >222 |
| Storage | Store at -20oC |
| Dissolution | Up to 50mM in DMSO |
| TP-422 |
 |
Click here to download the SDF file. |
SMILES:
CN(CCCOC1=CC=C(C2=NC(OC)=NC(OCCCN3C=CC=N3)=C2)C=C1)C
InChI:
InChI=1S/C22H29N5O3/c1-26(2)12-5-15-29-19-9-7-18(8-10-19)20-17-21(25-22(24-20)28-3)30-16-6-14-27-13-4-11-23-27/h4,7-11,13,17H,5-6,12,14-16H2,1-3H3
InChIKey:
CVRFBLOQBLKUBC-UHFFFAOYSA-N
| Physical and chemical properties | |
| Molecular weight | 411.22 |
| Molecular formula | C22 H29 N5 O3 |
| IUPAC name | 1-(3-(6-(4-(3-dimethylamino-propoxy)-phenyl)-2-methoxy-pyrimidin-4-yloxy)-propyl)-1H-pyrazole |
| clogP | 3.4 |
| PSA | 59.0 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 12 |
| No. of hydrogen bond acceptors | 7 |
| No. of hydrogen bond donors | 0 |
| PAMPA (nm/sec, pH=7.4) | 283 |
| Aqueous solubility (µM, pH= 6.8) | >267 |
| Storage | Store at -20oC |
| Dissolution | Up to 50mM in DMSO |
A DSF screen against Human bromodomains reveals only one significant off-target; BRD9:
ITC showed good potency and sufficient selectivity against BRD9 to meet SGC probe criteria:
Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Stratagene). Proteins were buffered in 10 mM HEPES pH 7.5, 500 mM NaCl and assayed in a 96-well plate at a final concentration of 2 μM in 20 μL volume. Compounds were added at a final concentration of 10 μM. SYPRO Orange (Molecular Probes) was added as a fluorescence probe at a dilution of 1:1000. Excitation and emission filters for the SYPRO-Orange dye were set to 465 nm and 590 nm, respectively. The temperature was raised with a step of 3 °C per minute from 25 °C to 96 °C and fluorescence readings were taken at each interval.
All bromodomain proteins were prepared according to the published procedures (Filippakopoulos at al, 2012). Assay was performed as described previously (Philpott et al, 2011). All reagents were pre-diluted in 25 mM HEPES, 100 mM NaCl, 0.1 % BSA, pH 7.4 and 0.05 % CHAPS and allowed to equilibrate to room temperature prior to addition to plates. Plates filled with 5 uL of the assay buffer followed by 7 uL of biotinylated peptide [H-YSGRGKacGGKacGLGKacGGAKacRHRK(Biotin)-OH and His-tagged protein to achieve final assay concentrations of 25 nM. Plates were sealed and incubated for a further 60 minutes, before the addition of 8 μl of the mixture of streptavidin-coated donor beads (12.5 μg/ml) and nickel chelate acceptor beads (12.5 μg/ml) under low light conditions. Plates were foil-sealed to protect from light, incubated at room temperature for 60 minutes and read on a PHERAstar FS plate reader (BMG Labtech, Germany) using an AlphaScreen 680 excitation/570 emission filter set.
Experiments were carried out on a VP-ITC microcalorimeter (MicroCal™). All experiments were performed at 15 °C in 25 mM HEPES pH 7.4, 150 mM NaCl, 500 μM TCEP. 50 mM stocks of compound was thawed and diluted in 2 mL of buffer to a final concentration of 10 µM in the ITC cell. The protein titrations were conducted using an initial injection of 2 µl followed by 30 identical injections of 6 µl. The dilution heats were measured on separate experiments and were subtracted from the titration data. Thermodynamic parameters were calculated using ∆G = ∆H - T∆S = -RTlnKB, where ∆G, ∆H and ∆S are the changes in free energy, enthalpy and entropy of binding respectively. In all cases a single binding site model was employed.
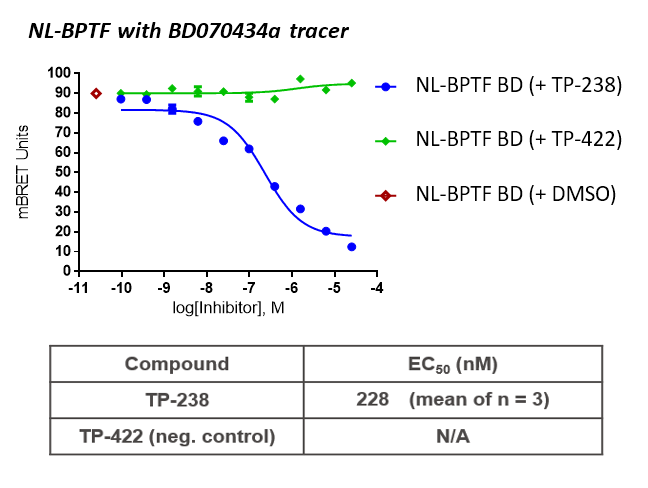
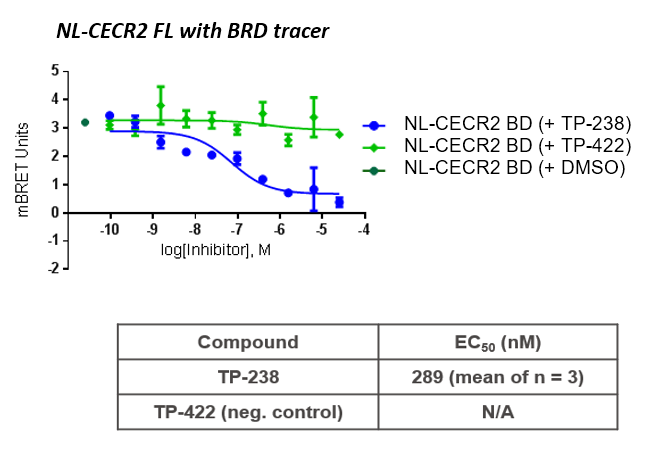
TP-238 and its negative control were tested for target engagement in HEK cells using NanoBRETTM. Significant inhibition by TP-238 against BPTF (n = 3, EC50 = 228 nM) and CECR2 (n = 3, EC50 = 289 nM) whereas the negative control TP-422 exhibited no significant activity at the tested concentrations:
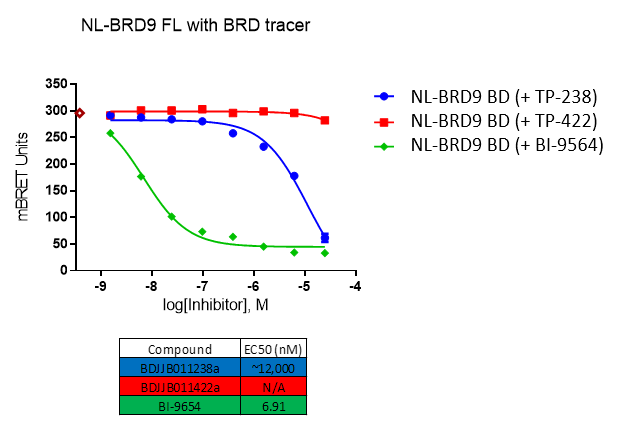
Further, no significant inhibition against BRD9 was found in a NanoBRETTM assay in HEK293 cells:
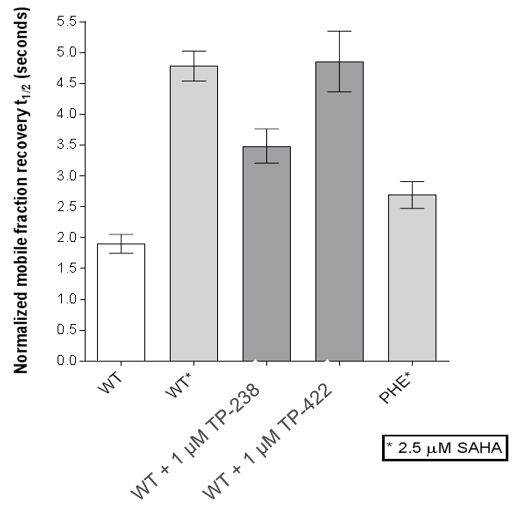
 FRAP assays were also performed which showed significant inhibition by TP-238 against BPTF and CECR2:
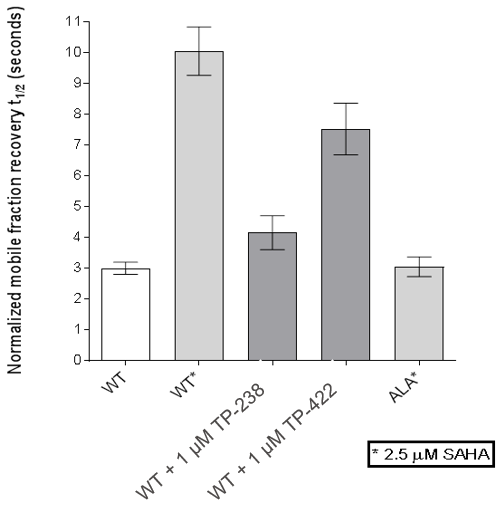
FRAP assays were also performed which showed significant inhibition by TP-238 against BPTF and CECR2:
FL-BPTF FRAP:
FL-CECR2 FRAP:
HEK cells were reverse transfected with NL-FL CECR2 or NL-BPTF BD. Cells were treated with a concentration range of TP-238, TP-422 (negative control), with BD070434a tracer (1 µM) for BPTF or BRD-02 tracer (0.5 µM) for CECR2 for 3 hrs before BRET measurements were taken.
For BRD9, the full length protein was used. HEK2983 cells were transfected with NL-BRD9 and treated with tracer (2 µM) and the respective compounds, followed by addition of Nano-Glo substrate and extracellular inhibitor. BRET was determined at 450 and 610nM.
FRAP assays were performed using U2OS cells reverse transfected with GFP-FL FALZ WT or #PHE or GFP-FL CECR2 WT or #ALA for 6-8 hrs. Transfection reagents were replaced with media or 2.5 µM SAHA + incubated O/N. Cells were treated with 1 µM test cpds for 1 hr before imaging. 6 repeat assays were performed for each protein.
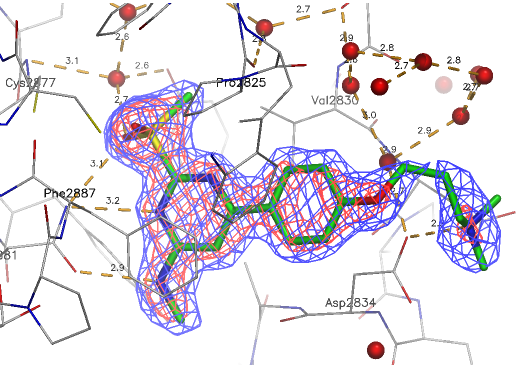
A close homologue to TP-248 was co-crystallised to investigate the possible binding mode of the probe:
| Probe | Negative control | |
 |  | |
VinSpinIn (Vinnie's Spindlin Inhibitor) | VinSpinIC (Vinnie's Spindlin Inactive Control) |
The Spindlin proteins are tudor domain containing proteins. There are five spindlin family members (Spin1, Spin2A, Spin2B, Spin3, Spin4), which are expressed at various levels throughout the body. However, spindlin1 (Spin1) is expressed at higher levels compared to the other family members.1 Our current knowledge of the biological roles of spindlins is limited to that of Spin1.
The Spin proteins consist of three tudor-like Spin/Ssty domains, arranged in a clockwise orientation.2-4 Spin1 has been shown to bind to trimethylated lysine 4 of histone 3 (H3K4me3) via domain 2 (ITC Kd = 147 nM).3,5 However, increased binding is observed when a second epigenetic methylation mark is present on arginine 8 (asymmetrically dimethylated) (H3K4me3R8me2a; ITC Kd = 45 nM).2 Spin1 also binds to H4K20me3 via domain 2.6
Spin1 binding to methylated histones is associated with transcriptional activation. Spin1 was found to be overexpressed in various cancers and has been shown to drive cancer cell proliferation through activation of the Wnt/β-catenin, PI3K/Akt and RET signalling pathways.2,7-10
Conversely, Spin1 has been shown to facilitate the inactivation of p53 by sequestering the ribosomal protein uL18.11
Direct or indirect Spin1 knockdown resulted in cancer cell and xenograft tumor growth inhibition, and such studies suggest that small molecule inhibition of Spin1 may be a viable approach for the treatment of certain cancers.8-13
Therefore, the SGC has developed VinSpinIn as a potent, cell active chemical probe for the Spin family proteins. VinSpinIn, along with the structurally very similar inactive control compound VinSpinIC, will contribute significantly to the elucidation of the biological roles and functions of the spindlin proteins, and will aid with the validation of Spin1 as a chemotherapeutic target.
Potency Against Target Family
The SYPRO Orange thermal shift assay was employed to assess the potency of VinSpinIn and VinSpinIC against four of the five Spin family members (Table 1). VinSpinIn induced a large shift in the thermal stability in all of the Spin protein assessed, while VinSpinIC did not. ITC was also performed to determine the potency of VinSpinIn on four Spin family members as well as an additional Spin1 construct (Table 1). VinSpinIn had KDs ranging between approximately 10-130 nM across the family.
| Spin Family Proteins | Thermal Shift Assay (ΔTm°C) | ITC Assay (KD nM) | |
| VinSpinIn | VinSpinIC | VinSpinIn | |
| Spin149-262 | Not tested | Not tested | 9.9 |
| SPIN126-262 | 13.17 | 1.02 | 111.1 |
| SPIN2B22-258 | 10.47 | 0.29 | 46.1 |
| SPIN321-258 | 14.12 | 2.34 | 131.1 |
| SPIN436-249 | 6.53 | 0.25 | 18.1 |
Table 1: Potency Against Target Family
Selectivity
VinSpinIn and VinSpinIC were screened against a panel of methyl binding domains (MBDs) using the thermal shift assay (Table 2). No significant thermal shift was observed for any of the MBDs screened.
A Scintillation Proximity Assay (SPA) was employed to screen VinSpinIn and VinSpinIC against a panel of methyltransferases and IC50s were determine for selected targets (Table 3). The lowest IC50 of VinSpinIn (PRMT4) was approximately 300 times greater than the AlphaScreen IC50 on Spin1 (30 nM).
| Methyl Lysine Binders | VinSpinIn (ΔTm°C) | VinSpinIC (ΔTm°C) |
| UHRF1139-298 | -0.04 | -0.14 |
| 53BP11483-1606 | -0.65 | -0.41 |
| TDRD3525-611 | 0.15 | -0.95 |
| SND1650-910 | -0.09 | -1.62 |
| SETDB1197-403 | -0.04 | -0.34 |
| SGF29129-293 | -0.05 | -0.92 |
| CCDC101114-293 | -0.04 | 0.3 |
| CHD1269-446 | -0.65 | 2.37 |
| FALZ2736-2793 | 0.15 | 0.31 |
| ING2211-266 | -0.09 | 0.25 |
| JARID1A1542-1660 | -0.04 | -0.04 |
| MLL1558-1773 | -0.05 | 0.63 |
| MLL5113-171 | -0.05 | -0.05 |
| PHF21-64 | 0.02 | 0.19 |
| PHF81-63 | 0.23 | -0.11 |
| TAF31086-1153 | -0.21 | -0.04 |
Table 2: Screening on a panel of MBDs.
| Methyltransferase Panel | VinSpinIn (IC50 µM) | VinSpinIC (IC50 µM) |
| PRMT4 | 9 | 2 |
| SETD2 | 20 | 5 |
| PRMT7 | 19 | 8 |
| SUV39H1 | 34 | 18 |
| PRMT6 | 27 | 19 |
| PRC2 | 21 | 27 |
| SMYD2 | 22 | 44 |
| PRDM9 | 25 | 44 |
| PRMT1 | 47 | 59 |
| PRMT8 | 45 | 66 |
Table 3: IC50s on methyltransferases.
Dosage
Use between 0.5 and 3 µM for Cellular Assays and 1 µM for screening at a single shot, for both VinSpinIn & VinSpinIC.
In vitro Activity
In a number of biophysical assays, VinSpinIn was shown to be a potent Spin1 inhibitor and has an ITC KD which is approximately 130 times more potent than that of the inactive control VinSpinIC (Table 4).
| Compound | AlphaScreen (Spin126-262) (IC50) | Octet BLI (Spin126-262) (KD) | ITC (Spin126-262) (KD) | ITC (Spin149-262) (KD) | Tm Shift (Spin126-262) (ΔTm°C) |
| VinSpinIn | 30 nM | 55 nM | 111.1 nM | 9.9 nM | 13.2 |
| VinSpinIC | 3.64 µM | not tested | not tested | 1.3 µM | 1.0 |
Table 4: In vitro biophysical binding assay results of VinSpinIn & VinSpinIC.
Cellular Activity
In a NanoBRET cellular target engagement assay VinSpinIn displayed dose dependant inhibition of the Spin1-H3 interaction; the inactive VinSpinIC showed no inhibition (Figure 2).
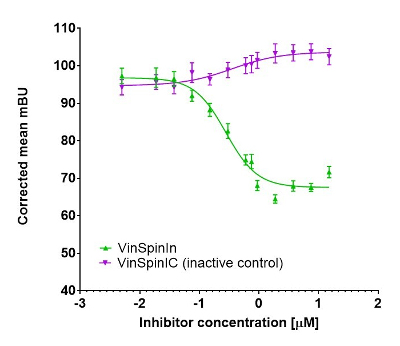
Figure 2: NanoBRET cellular engagement assay. Click on the ‘Cell-based Assay Data’ tab above for more details.
| VinSpinIn |
 |
Click here to download the SDF file. |
| VinSpinIn | |
| Physical and chemical properties | |
| Molecular weight | 738.96 g/mol |
| Molecular formula | C42H58N8O4 |
| IUPAC name | 2-[4-(2-{[2-(3-{[2-amino-5-(cyclopropylmethoxy)-3,3-dimethyl-3H-indol-6-yl]oxy}propyl)-2,3-dihydro-1H-isoindol-5-yl]oxy}ethyl)-1H-1,2,3-triazol-1-yl]-1-{4-[2-(pyrrolidin-1-yl)ethyl]piperidin-1-yl}ethan-1-one |
| logP | 3.85 |
| TPSA | 123.57 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 17 |
| No. of hydrogen bond acceptors | 2 |
| No. of hydrogen bond donors | 12 |
| Storage | +4 °C |
| Dissolution | DMSO (up to at least 50 mM) |
| VinSpinIC |
 |
Click here to download the SDF file. |
| VinSpinIC | |
| Physical and chemical properties | |
| Molecular weight | 738.96 g/mol |
| Molecular formula | C42H58N8O4 |
| IUPAC name | 2-[4-({[2-(4-{[2-amino-5-(cyclopropylmethoxy)-3,3-dimethyl-3H-indol-6-yl]oxy}butyl)-2,3-dihydro-1H-isoindol-5-yl]oxy}methyl)-1H-1,2,3-triazol-1-yl]-1-{4-[2-(pyrrolidin-1-yl)ethyl]piperidin-1-yl}ethan-1-one |
| clogP | 3.99 |
| TPSA | 123.57 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 17 |
| No. of hydrogen bond acceptors | 2 |
| No. of hydrogen bond donors | 12 |
| Storage | +4 °C |
| Dissolution | DMSO (up to at least 50 mM) |
SMILES:
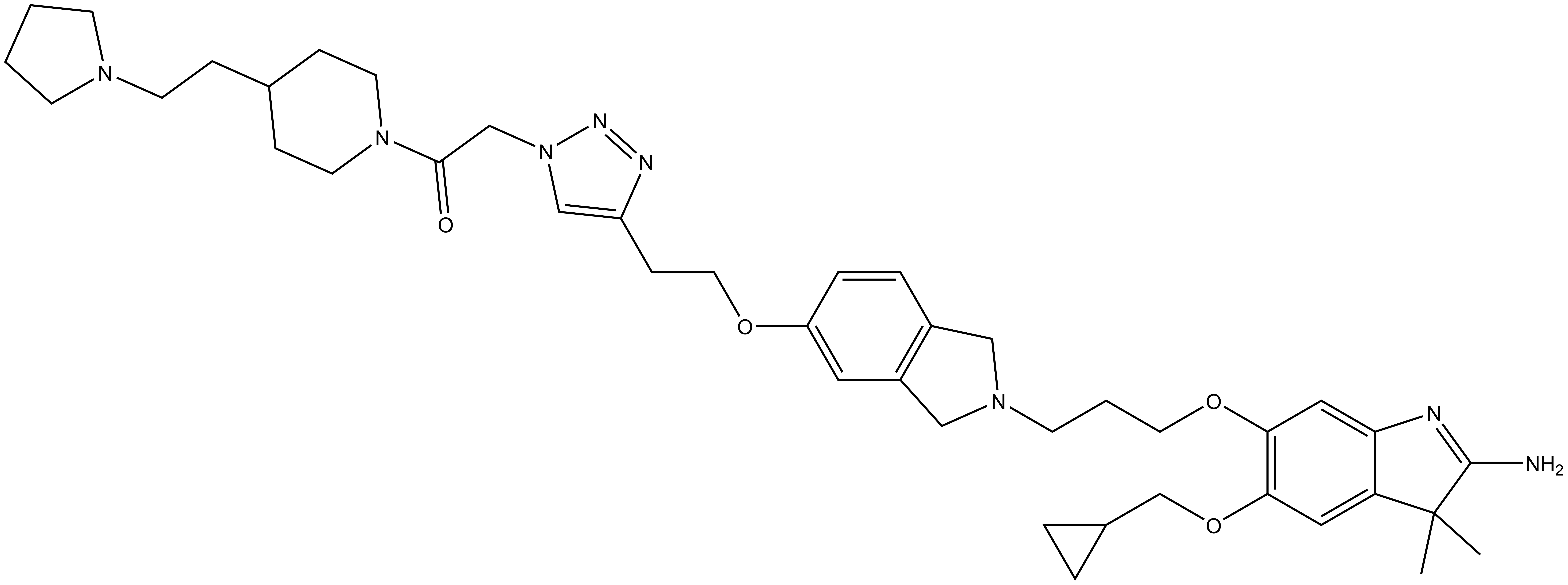
VinSpinIn: CC1(C(N)=NC2=CC(OCCCN3CC4=C(C3)C=C(OCCC5=CN(N=N5)CC(N6CCC(CC6)CCN7CCCC7)=O)C=C4)=C(C=C21)OCC8CC8)C
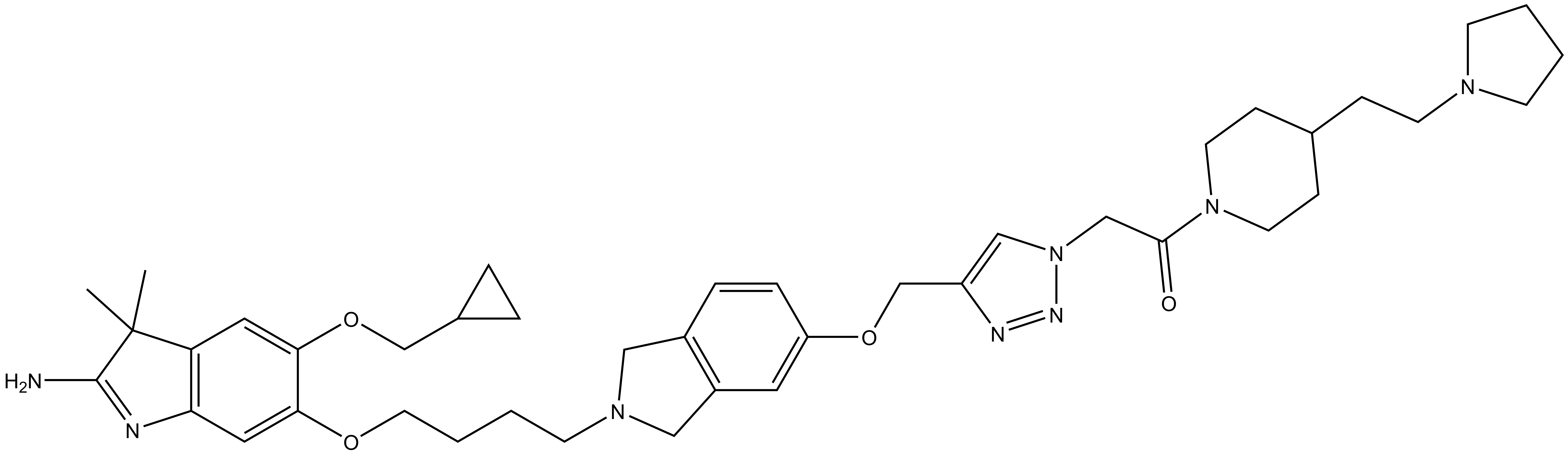
VinSpinIC: O=C(CN1N=NC(COC2=CC3=C(C=C2)CN(C3)CCCCOC4=CC5=C(C(C)(C(N)=N5)C)C=C4OCC6CC6)=C1)N7CCC(CC7)CCN8CCCC8
InChI:
VinSpinIn: InChI=1/C42H58N8O4/c1-42(2)36-23-38(54-29-31-6-7-31)39(24-37(36)44-41(42)43)53-20-5-16-48-25-32-8-9-35(22-33(32)26-48)52-21-13-34-27-50(46-45-34)28-40(51)49-18-11-30(12-19-49)10-17-47-14-3-4-15-47/h8-9,22-24,27,30-31H,3-7,10-21,25-26,28-29H2,1-2H3,(H2,43,44)/f/h43H2
VinSpinIC: InChI=1/C42H58N8O4/c1-42(2)36-22-38(54-28-31-7-8-31)39(23-37(36)44-41(42)43)52-20-6-5-16-48-24-32-9-10-35(21-33(32)25-48)53-29-34-26-50(46-45-34)27-40(51)49-18-12-30(13-19-49)11-17-47-14-3-4-15-47/h9-10,21-23,26,30-31H,3-8,11-20,24-25,27-29H2,1-2H3,(H2,43,44)/f/h43H2
InChIKey:
VinSpinIn: XPEJZXWPKDAYFX-UHFFFAOYSA-N
VinSpinIC: FOBGCBSESHUBEI-UHFFFAOYSA-N
The SYPRO Orange thermal shift assay was employed to screen VinSpinIn and VinSpinIC against a panel of methyl binding domains (MBDs), which included four of the five Spin family members (Table 1). VinSpinIn induced a large shift in thermal stability of Spin1, Spin2B, Spin3, Spin4. No other significant shift in thermal stability was observed for VinSpinIn or the inactive VinSpinIC.
| Methyl Lysine Binders | VinSpinIn (ΔTm°C) | VinSpinIC (ΔTm°C) |
| a UHRF1139-298 | -0.04 | -0.14 |
| a 53BP11483-1606 | -0.65 | -0.41 |
| a TDRD3525-611 | 0.15 | -0.95 |
| a SND1650-910 | -0.09 | -1.62 |
| a SETDB1197-403 | -0.04 | -0.34 |
| a SGF29129-293 | -0.05 | -0.92 |
| b CCDC101114-293 | -0.04 | 0.3 |
| b CHD1269-446 | -0.65 | 2.37 |
| b FALZ2736-2793 | 0.15 | 0.31 |
| b ING2211-266 | -0.09 | 0.25 |
| b JARID1A1542-1660 | -0.04 | -0.04 |
| b MLL1558-1773 | -0.05 | 0.63 |
| b MLL5113-171 | -0.05 | -0.05 |
| b PHF21-64 | 0.02 | 0.19 |
| b PHF81-63 | 0.23 | -0.11 |
| b TAF31086-1153 | -0.21 | -0.04 |
| b SPIN126-262 | 13.17 | 1.02 |
| b SPIN2B22-258 | 10.47 | 0.29 |
| b,c SPIN321-258 | 14.12 | 2.34 |
| b SPIN436-249 | 6.53 | 0.25 |
Table 1: Screening on a panel of MBDs.a protein = 0.05 to 0.2 mg/ml, compound = 200 µM; b protein = 2 µM, compound = 20 µM; c Intrinsic tryptophan fluorescence used
A fluorescence polarization displacement assays was employed to screen the compounds against L3MBTL1 and L3MBTL3 (MBT methyl lysine readers), which gave respective Kdisp.s of 27 and 8 µM for VinSpinIn, and 28 and 14 µM for the inactive VinSpinIC. Therefore, VinSpinIn is approximately 267 times less potent towards L3MBTL1 than Spin1.
Selectivity screening was also performed against a panel of methyl transferases domains (MTDs) including protein, DNA and RNA methyltransferases using a Scintillation Proximity Assay (SPA) (Figure 1).
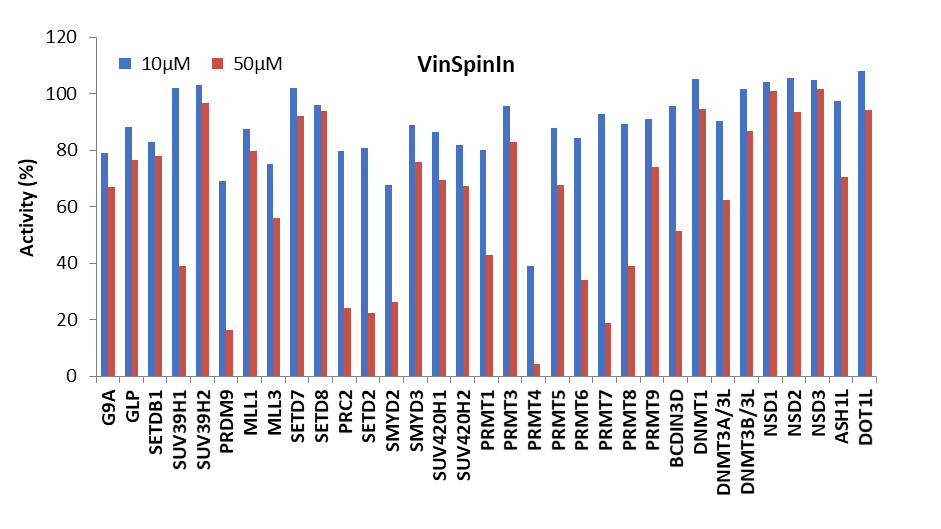
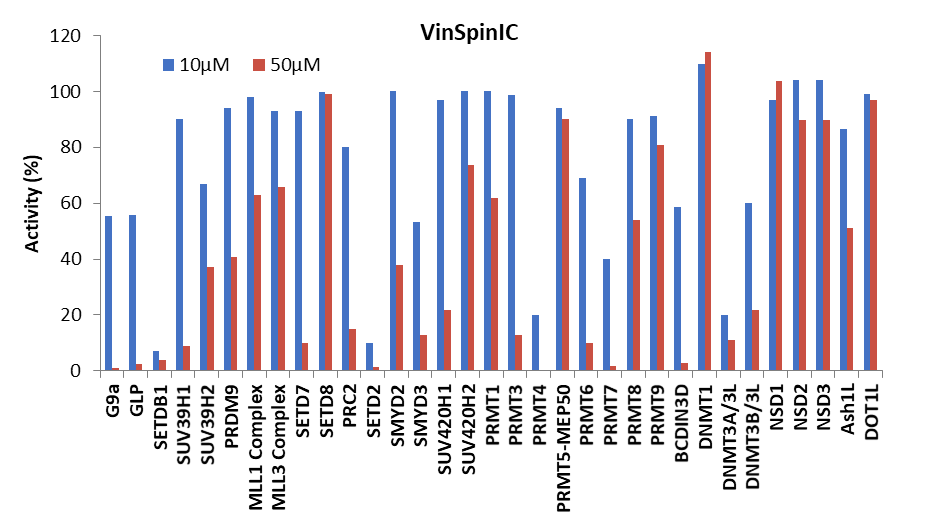
Figure 1: (Top) Methyltransferase activity in presence of VinSpinIn at 10 & 50 µM
(Bottom) Methyltransferase activity in presence of VinSpinIC at 10 and 50 µM.
For selected targets IC50s were determined (Table 2). The lowest IC50 of VinSpinIn (PRMT4) was approximately 300 times greater than the AlphaScreen IC50 on Spin1.
| Methyltransferase Panel | VinSpinIn (IC50 µM) | VinSpinIC (IC50 µM) |
| PRMT4 | 9 | 2 |
| SETD2 | 20 | 5 |
| PRMT7 | 19 | 8 |
| SUV39H1 | 34 | 18 |
| PRMT6 | 27 | 19 |
| PRC2 | 21 | 27 |
| SMYD2 | 22 | 44 |
| PRDM9 | 25 | 44 |
| PRMT1 | 47 | 59 |
| PRMT8 | 45 | 66 |
| DNMT3A/3L | a | 9 |
| DNMT3B/3L | a | 5 |
| G9a | a | 9 |
| GLP | a | 21 |
| SETDB1 | a | 6 |
Table 2: IC50s of selected methyltransferases.a Not determined
Effects of VinSpinIn and VinSpinIC on methyltransferase activity of G9a, GLP, SUV39H1, SUV39H2, SETDB1, SETD8, SUV420H1, SUV420H2, SETD7, MLL1 trimeric complex, MLL3 pentameric complex, EZH2 trimeric complex, PRMT1, PRMT3, PRMT4, PRMT5-MEP50 complex, PRMT6, PRMT7, PRMT8, PRMT9, PRDM9, SETD2, SMYD2, SMYD3, and DNMT1 was assessed by monitoring the incorporation of tritium-labeled methyl group to lysine or arginine residues of peptide substrates using Scintillation Proximity Assay (SPA) as previously described14. Assays were performed in a 10 µl reaction mixture containing 3H-SAM (Cat.# NET155V250UC; Perkin Elmer; www.perkinelmer.com) at substrate concentrations close to Km values for each enzyme. Two concentrations (10µM and 50 µM) of VinSpinIn or VinSpinIC were used in all selectivity assays. To stop the enzymatic reactions, 10 µl of 7.5 M guanidine hydrochloride was added, followed by 180 µl of buffer (20 mM Tris, pH 8.0), mixed and then transferred to a FlashPlate (Cat.# SMP103; Perkin Elmer; www.perkinelmer.com). After mixing, the reaction mixtures in Flash plates were incubated for 2 hours and the CPM were measured using Topcount plate reader (Perkin Elmer, www.perkinelmer.com). The CPM counts in the absence of compound for each data set were defined as 100% activity. In the absence of the enzyme, the CPM counts in each data set were defined as background (0%).
For DOT1L, NSD1, NSD2, NSD3, ASH1L, DNMT3A/3L, and DNMT3B/3L, a filter-based assay was used. In this assay, 10 µl of reaction mixtures were incubated at 23 oC for 1 hour, 50 µl of 10% trichloroacetic acid (TCA) was added, mixed and transferred to filter-plates (Millipore; cat.# MSFBN6B10; www.millipore.com). Plates were centrifuged at 2000 rpm (Allegra X-15R - Beckman Coulter, Inc.) for 2 min followed by 2 additional 10% TCA wash and one ethanol wash followed by centrifugation. Plates were dried and 30 µl MicroO (MicroScint-O; Cat.# 6013611, Perkin Elmer; www.perkinelmer.com) was added to each well, centrifuged and removed. 50 µl of MicroO was added again and CPM was measured using Topcount plate reader.
IC50 Determinations:
IC50 values were determined for inhibition of methyltransferase activity of the following enzymes:
To stop the enzymatic reactions, 7.5 M Guanidine hydrochloride was added, followed by 180 µL of buffer (20 mM Tris, pH 8.0), mixed and then transferred to a FlashPlate (Cat.# SMP103; Perkin Elmer; www.perkinelmer.com). After mixing, the reaction mixtures in Flash plate were incubated for 2 hour and the CPM counts were measured using Topcount plate reader (Perkin Elmer, www.perkinelmer.com). The CPM counts in the absence of compound for each data set were defined as 100% activity. In the absence of the enzyme, the CPM counts in each data set were defined as background (0%). The IC50 values were calculated using GraphPad Prism 7 software.
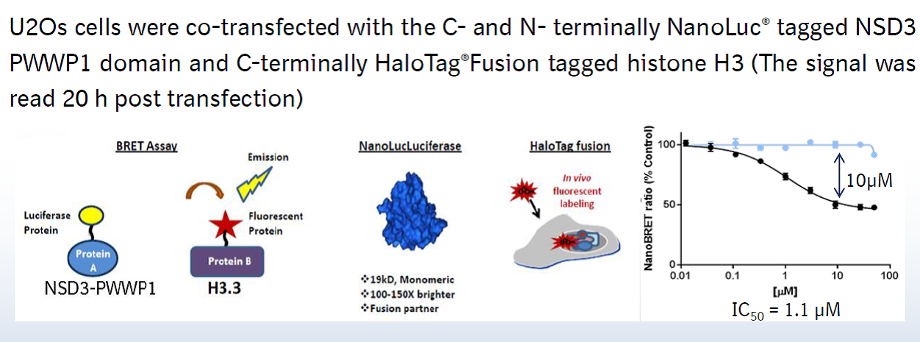
In a NanoBRET cellular target engagement assay VinSpinIn displayed dose dependant inhibition of the Spin1-H3 interaction, with an IC50 of 270 nM. The inactive VinSpinIC showed no inhibition (Figure 1).
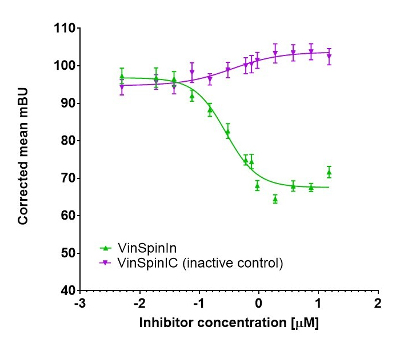
Figure 1: NanoBRET cellular target engagement assay performed in U2OS cells. Full length SPIN1 with N-terminal NanoLuc; Histone 3.3 with C-terminal Halotag; 24h incubation with VinSpinIn and VinSpinIC
U20S cell (2.8 x 105) were plated in each well of a 6-well plate after 6h cells were co-transfected with C-terminal HaloTag-Histone 3.3 (NM_002107) and an N-terminal NanoLuciferase fusion of full length SPIN1 at a 1:500 (NanoLuc® to HaloTag®) ratio respectively with FuGENE HD transfection regent15.
Sixteen hours post-transfection, cells were collected, washed with PBS, and exchanged into media containing phenol red-free DMEM and 4% FBS in the absence (control sample) or the presence (experimental sample) of 100 nM NanoBRET 618 fluorescent ligand (Promega). Cells were then re-plated in a 384-well assay white plate (Greiner #3570) at 2.7x103 cells per well. VinSpinIn and VinSpinIC were then added directly to media at final concentrations 0-30μM or an equivalent amount of DMSO as a vehicle control, and the plates were incubated for 24 h at 37oC in the presence of 5% CO2.
NanoBRET Nano-Glo substrate (Promega) was added to both control and experimental samples at a final concentration of 10 µM. Readings were performed within 10 minutes using a ClarioSTAR (BMG labtech) equipped with 460 nm and 610 nm filters. A corrected BRET ratio was calculated and is defined as the ratio of the emission at 610 nm/460 nm for experimental samples minus the emission at 610 nm/460 nm for control samples (without NanoBRET fluorescent ligand). BRET ratios are expressed as milliBRET units (mBU), where 1 mBU corresponds to the corrected BRET ratio multiplied by 1000.
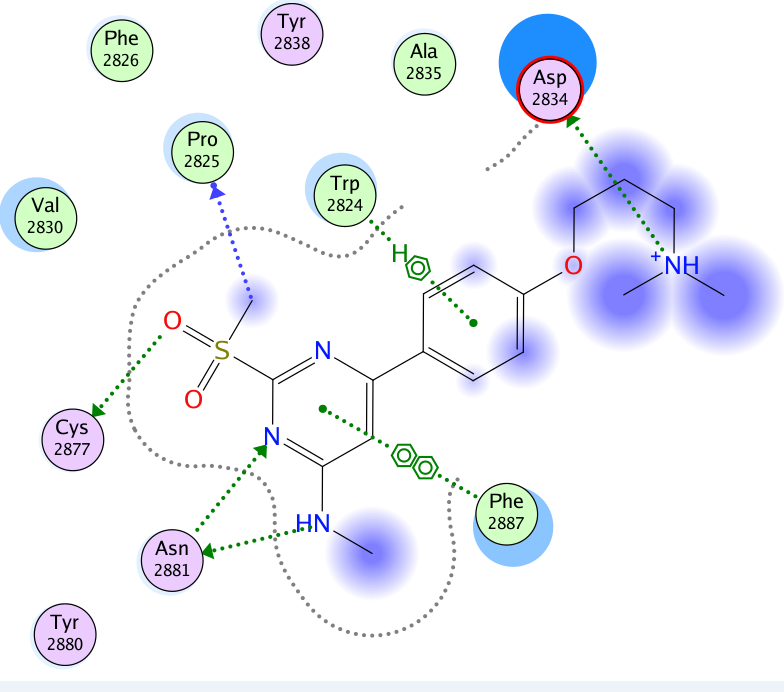
The co-crystal structure of Spin1 with VinSpinIn confirms that VinSpinIn binds to both domain 1 and 2 of Spin1, and therefore is referred to as a bidentate inhibitor (Figure 1).
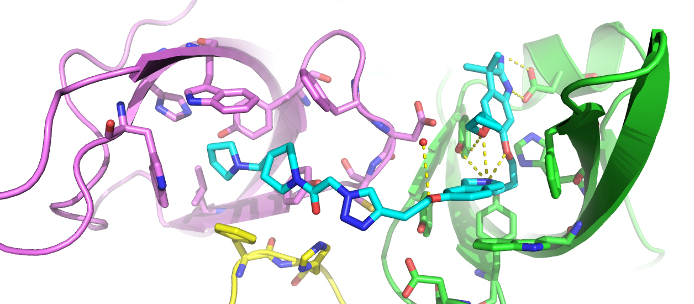
Figure 1: Co-crystal structure of Spin1 (domain 1 = purple; domain 2 = green; domain 3 = yellow) with VinSpinIn (Cyan).
The probe DDR-TRK-1 is available from Cayman Chemical and Sigma.
| Probe | Negative control | |
 |
|  |
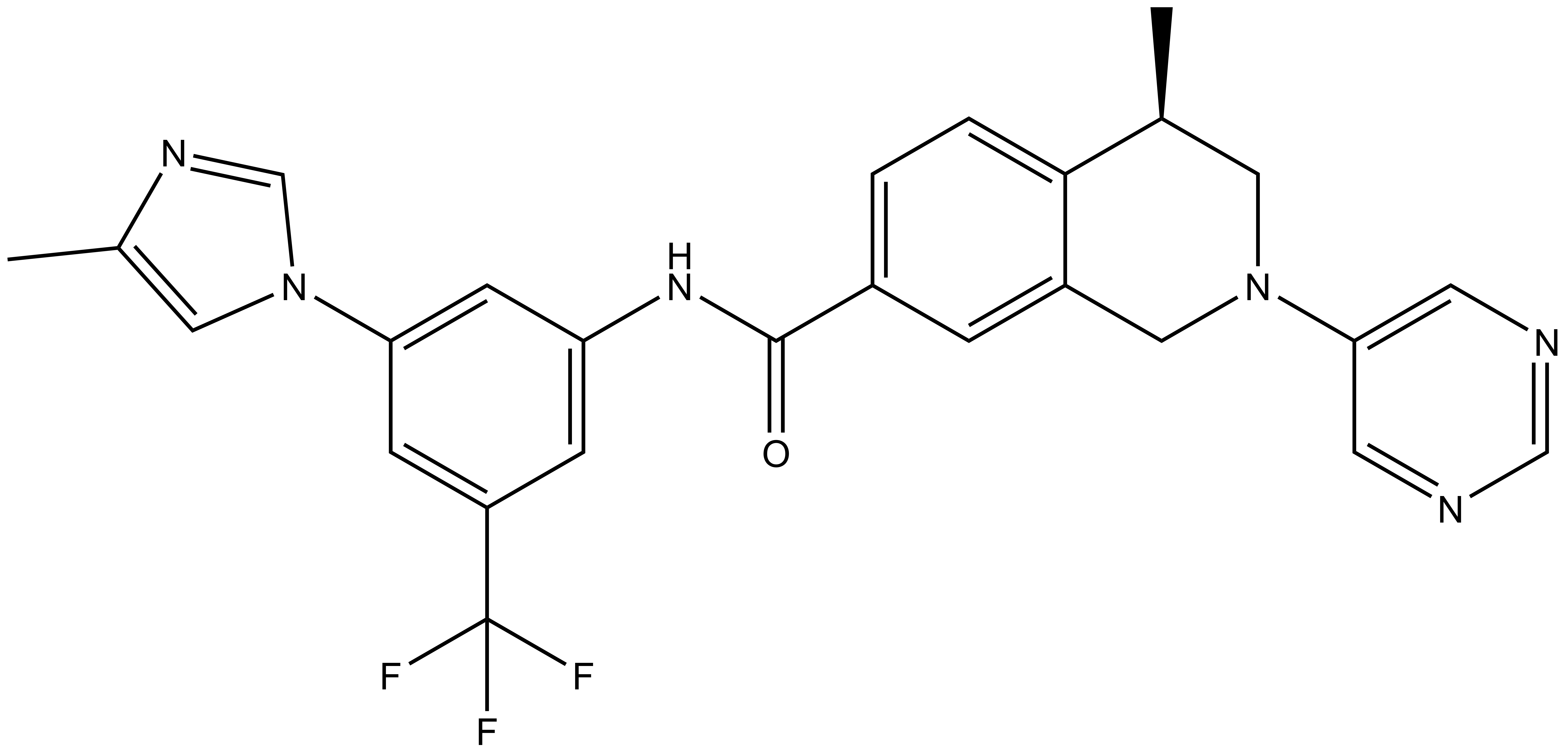
DDR-TRK-1 |
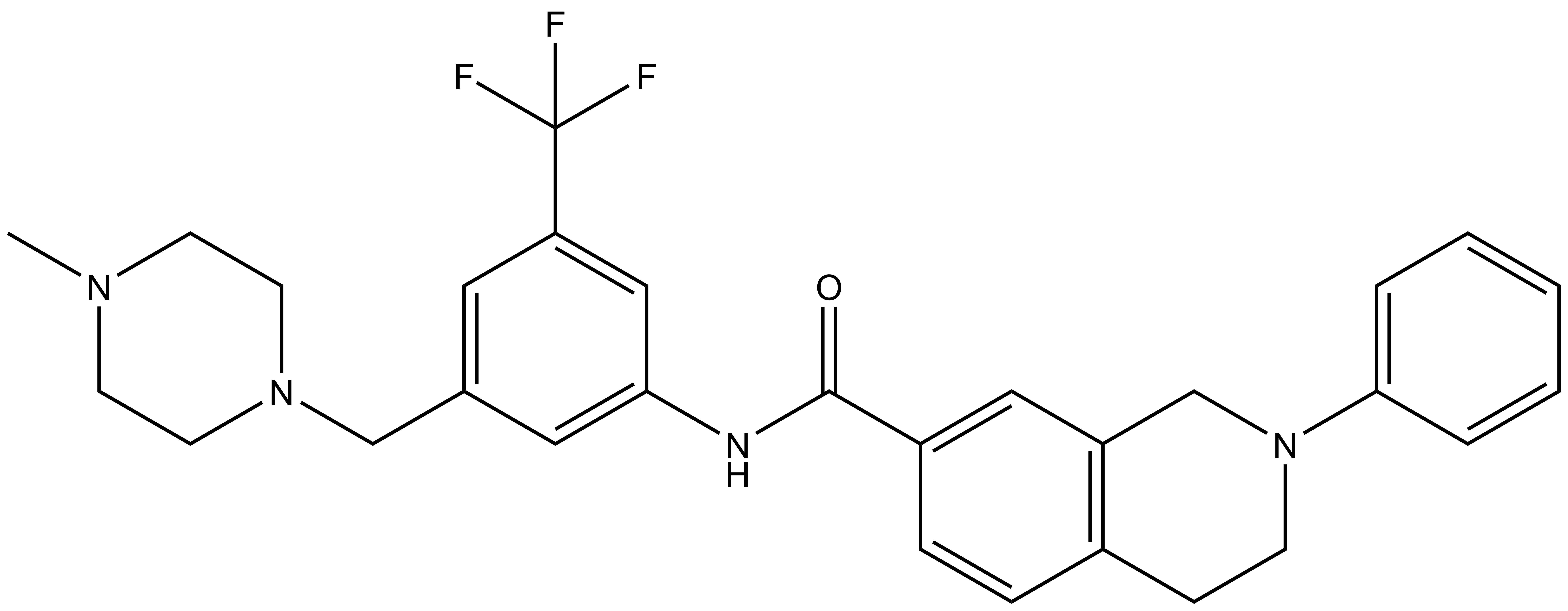
| DDR-TRK-1N |
The discoidin domain receptors (DDRs), DDR1 and DDR2, are unique among the receptor tyrosine kinases (RTKs) in being activated by interaction with the extracellular matrix via binding to triple-helical collagen by the receptor extracellular domains.1 DDR1 and DDR2 form constitutive dimers making them unusual among RTKs, which typically dimerize only upon activation.2 DDRs regulate extracellular matrix remodeling, as well as cell adhesion, proliferation and migration.3 DDR kinases are linked to the progression of various human diseases, including fibrotic disorders, atherosclerosis and cancer.3-5 Significantly, they are identified as indicators of poor prognosis in ovarian, breast and lung cancer.6 DDR1 overexpression is associated with increased cell survival and invasion in hepatocellular carcinomas, pituitary adenoma and prostate cancer,7 whereas DDR2 is mutated in squamous cell lung cancers8 and contributes to breast cancer metastasis.9 The promise of DDR kinases as a therapeutic target has been demonstrated by DDR1 knockdown that has been shown to reduce metastatic activity in lung cancer models,10 slow the development of atherosclerosis,5 and impede the development of fibrotic disorders.11
The TRK kinases are represented by three members, TRKA, TRKB, and TRKC, which are selectively expressed in neuronal tissue. Receptor signaling is initiated by binding of the neurotrophic factors NGF, BDNF, and NTR, respectively. Subsequent signaling is via the RAS, PLCγ and PI3γ pathways. The TRKs play a vital role in CNS development and survival. Gene fusions, protein overexpression, and single nucleotide alterations, have been implicated in the pathogenesis of specific cancer types including glioblastoma, papillary thyroid carcinoma, and secretory breast carcinomas, but are rare in most other cancers.12
DDR-TRK-1 is a chemical probe for the DDR and TRK kinases with good in vitro and in vivo potency.
DDR-TRK-1 inhibits colony formation and migration of Panc-1 pancreatic cancer cells. In cellular and mouse models of lung fibrosis, DDR-TRK-1 inhibits signaling, expression of fibrotic markers and fibrotic features such as hydroxproline expression. With restricted CNS exposure and therefore no TRK inhibition, the in vivo effects of DDR-TRK-1 can be attributed to DDR1-2 inhibition. DDR-TRK-1N is the negative control compound with very minimal differences in compound structure, and should be used in parallel to DDR-TRK-1. The probe can be complemented by the use of BAY-826, which inhibits the kinases TIE1, TIE2, DDR1 and DDR2, but lacks TRK activity to better understand the target involved in the phenotypic effect.
Work on this probe has been published in J.Med.Chem. 2016 59(12), p 5911-5916, 'Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) inhibitors'.
DDR-TRK-1 is a chemical probe for the DDR and TRK kinases (IC50 3-43 nM) with corresponding good cellular potency in NanoBRETTM target engagement assays (IC50 104-448 nM). DDR-TRK-1 was shown to be selective in an in vitro kinase panel followed by cellular NanoBRETTM assays.
DDR-TRK-1 is selective in KINOMEscan® at 1μM. The closest off target is CDK11 (370 nM), which was however shown to be only poorly inhibited in cells (~5 µM in NanoBRETTM). The negative control DDR-TRK-1N is entirely clean in KINOMEscan at 1μM.
We recommend that DDR-TRK-1 be used at 5μM concentration in cells. The negative control DDR-TRK-1N should be used at 5μM concentration in cells – it is toxic in HeLa cells above 10μM.
We also recommend the use of selective TRK inhibitors or the TIE-DDR BAY-826 probe in parallel to dissect the biology of DDR1/2 versus TRKA/B/C.
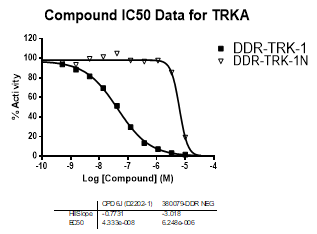
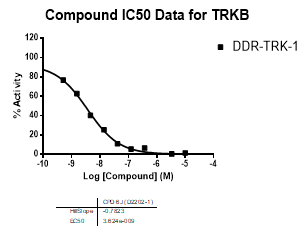
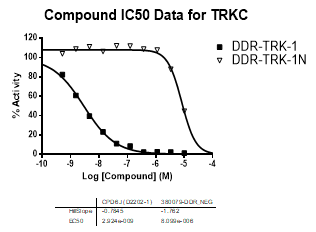
In NanoBRETTM assays, DDR-TRK-1 shows a potency of 104nM against DDR1, 175nM against DDR2, 448nM against TRKA and 142nM against TRKB.
In an activity assay at RBC (10μM ATP), DDR-TRK-1 shows an IC50 value of 27nM against DDR1, 4.5nM against DDR2, 43nM against TRKA, 3.6nM against TRKB and 2.9nM against TRKC.
| Probe | Negative control | |
 |
|  |
DDR-TRK-1 |
| DDR-TRK-1N |
| Click here to download the DDR-TRK-1 SDF file. | Click here to download the DDR-TRK-1N SDF file. |
| Physical and chemical properties for DDR-TRK1 | |
| Molecular weight | 492.1885 |
| Molecular formula | C29 H31 F3 N4 O |
| IUPAC name | (3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoro-methyl)-phenylamino)-(5-methyl-3-(pyrimidin-5-yl)-3-aza-bicyclo[4.4.0]deca-1(6),7,9-trien-9-yl)-methanone |
| MollogP | 4.6 |
| PSA | 59.2 |
| No. of chiral centres | 1 |
| No. of rotatable bonds | 6 |
| No. of hydrogen bond acceptors | 5 |
| No. of hydrogen bond donors | 1 |
| Storage | -20 as DMSO stock |
| Dissolution | Soluble in DMSO at least up to 50mM |
| Physical and chemical properties for DDR-TRK-1N | |
| Molecular weight | 508.2450 |
| Molecular formula | C26 H23 F3 N6 O |
| IUPAC name | (3-((4-methyl-piperazin-1-yl)-methyl)-5-(trifluoro-methyl)-phenylamino)-(3-phenyl-3-aza-bicyclo[4.4.0]deca-1(6),7,9-trien-9-yl)-methanone |
| MollogP | 5.6 |
| PSA | 33.6531 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 7 |
| No. of hydrogen bond acceptors | 4 |
| No. of hydrogen bond donors | 1 |
| Storage | -20 as DMSO stock |
| Dissolution | Soluble in DMSO at least up to 50mM |
SMILES:
DDR-TRK-1: CC1=CN(C2=CC(NC(C3=CC=C4C(CN(C5=CN=CN=C5)C[C@@H]4C)=C3)=O)=CC(C(F)(F)F)=C2)C=N1
DDR-TRK-1N: CN1CCN(CC2=CC(C(F)(F)F)=CC(NC(C3=CC4=C(CCN(C5=CC=CC=C5)C4)C=C3)=O)=C2)CC1
InChI:
DDR-TRK-1: InChI=1S/C26H23F3N6O/c1-16-11-34(23-9-30-14-31-10-23)13-19-5-18(3-4-24(16)19)25(36)33-21-6-20(26(27,28)29)7-22(8-21)35-12-17(2)32-15-35/h3-10,12,14-16H,11,13H2,1-2H3,(H,33,36)/t16-/m0/s1
DDR-TRK-1N: InChI=1S/C29H31F3N4O/c1-34-11-13-35(14-12-34)19-21-15-25(29(30,31)32)18-26(16-21)33-28(37)23-8-7-22-9-10-36(20-24(22)17-23)27-5-3-2-4-6-27/h2-8,15-18H,9-14,19-20H2,1H3,(H,33,37)
InChIKey:
DDR-TRK-1: CMJJZRAAQMUAFH-INIZCTEOSA-N
DDR-TRK-1N: FWKRCZKMCJOFNG-UHFFFAOYSA-N
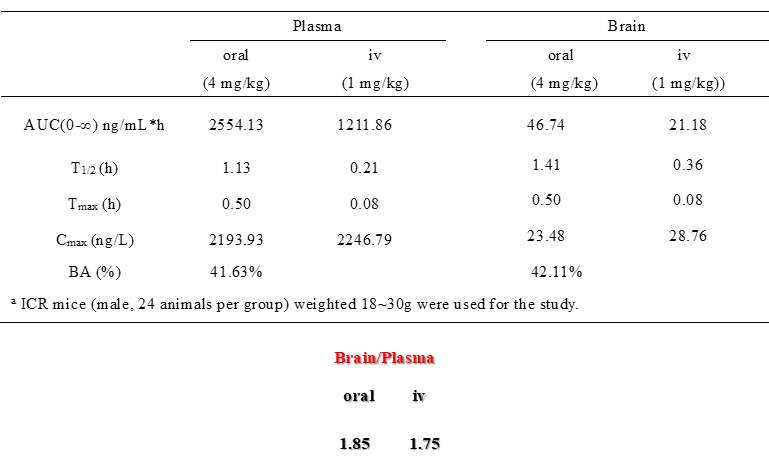

A KINOMEscan of DDR-TRK-1 at 1μM revealed very few off-targets:
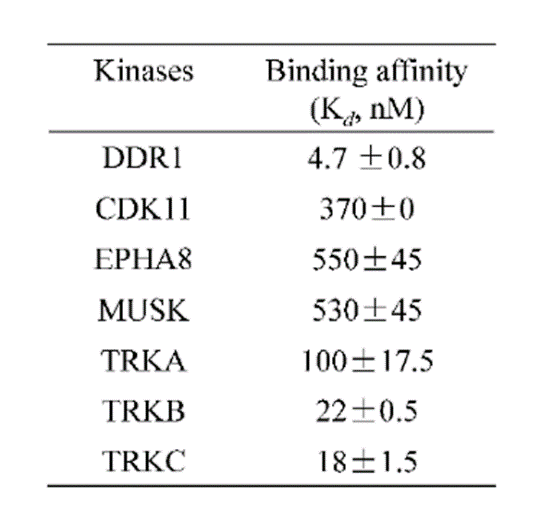
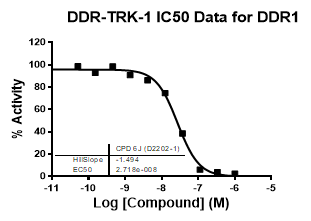
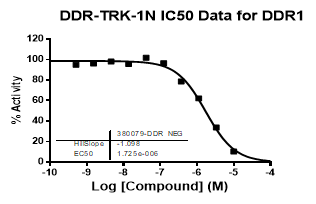
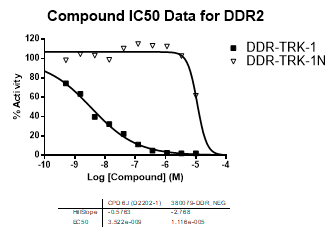
An activity assay at RBC (10μM ATP) revealed the following within the DDR/TRK family:
| Compound IC50 (M): | ||||
| Kinase: | DDR-TRK-1 | DDR-TRK-1N | Window probe/control | |
| DDR1 | 2.72E-08 | 1.73E-06 | 24 | |
| DDR2 | 3.52E-09 | >1.00E-05 | NA | |
| TRKA | 4.33E-08 | 6.25E-06 | 144 | |
| TRKB | 3.62E-09 | NA | ||
| TRKC | 2.92E-09 | 8.10E-06 | >2000 | |
| Dose-response curves for the probe (and where collected, the negative control) are shown below: |
 |
 |
 |
 |
 |
 |
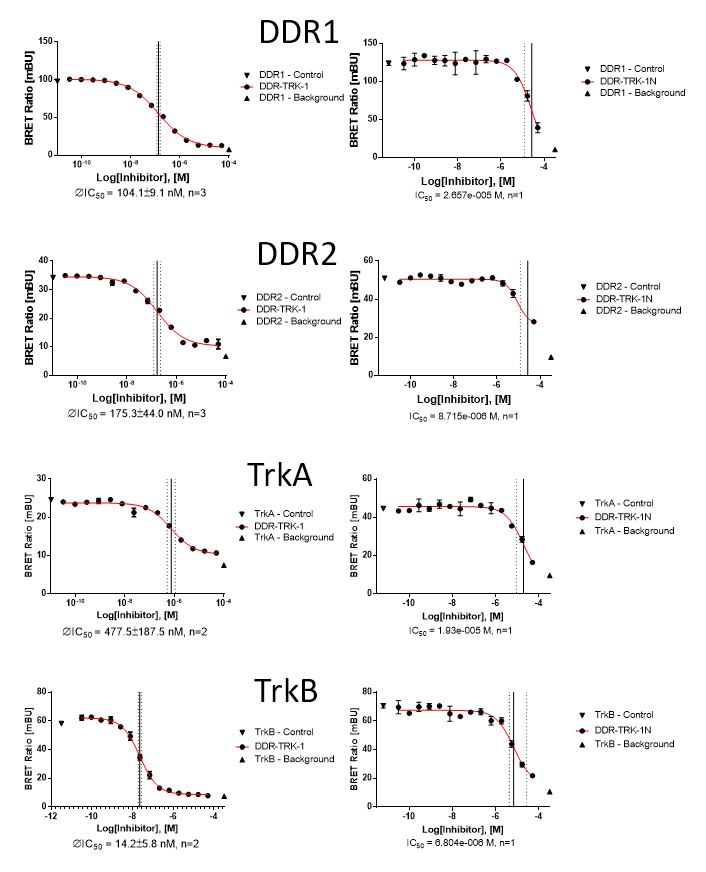
In a NanoBRETTM cellular target engagement assay DDR1-TRK-1 displayed dose dependant inhibition as follows:
| DDR-TRK-1 (M) | DDR-TRK-1N (M) | Window | |
| DDR1 | 1.04E-07 | 2.66E-05 | 256 |
| DDR2 | 1.75E-07 | 8.72E-06 | 50 |
| TRKA | 4.48E-07 | 1.93E-05 | 43 |
| TRKB | 1.42E-08 | 6.80E-06 | 478 |
NanoBRET assay
Dose-response experiments were conducted in 96 well format using HEK293T cells expressing NanoLuc fused to the C-terminus of full-length protein kinases for DDR2, TRKA and TRKB or isoform 2 for DDR1, using Promega tracer as indicated below.
| Kinase | Nluc location | full-length? | Tracer | [Tracer], nM (@Tracer) |
| DDR1 | C | Isoform 2 (Uniprot Q08345-2) | 4 | 85 |
| DDR2 | C | canonical (Uniprot Q16832-1) | 4 | 45 |
| TrkA | C | canonical (Uniprot P04629-1) | 5 | 50 |
| TrkB | C | canonical (Uniprot Q16620-1) | 5 | 30 |
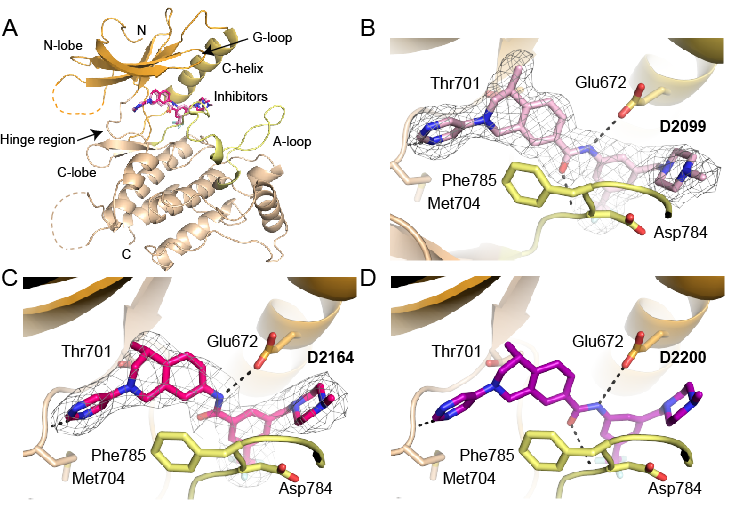
Structures of DDR1 complexes. A. Overall view of DDR1. Structural features are labelled for clarity. N-lobe is coloured orange, A loop is marked in yellow and C-lobe is coloured wheat. B. D2099 compound electron density (PDB 5FDP). C. D2164 compound electron density (PDB 5FDX). D. Molecular docking of DDR-TRK-1 compound.
Inhibition of DDR1, DDR2, TRKA, TRKB and TRKC kinase activity was measured using radiometric assay (Reaction Biology Corporation). Compounds were tested in 10-dose IC50 singlicate mode with a 3-fold serial dilution starting at 1 or 10 μM at [ATP] = 10 µM.
Kinome-wide profiling was performed at DiscoverX using the KINOMEscan assay.
The probe and control are no longer available.
| Probe | Negative control | |
 |
|  |
SKI-73 |
| SKI-73N |
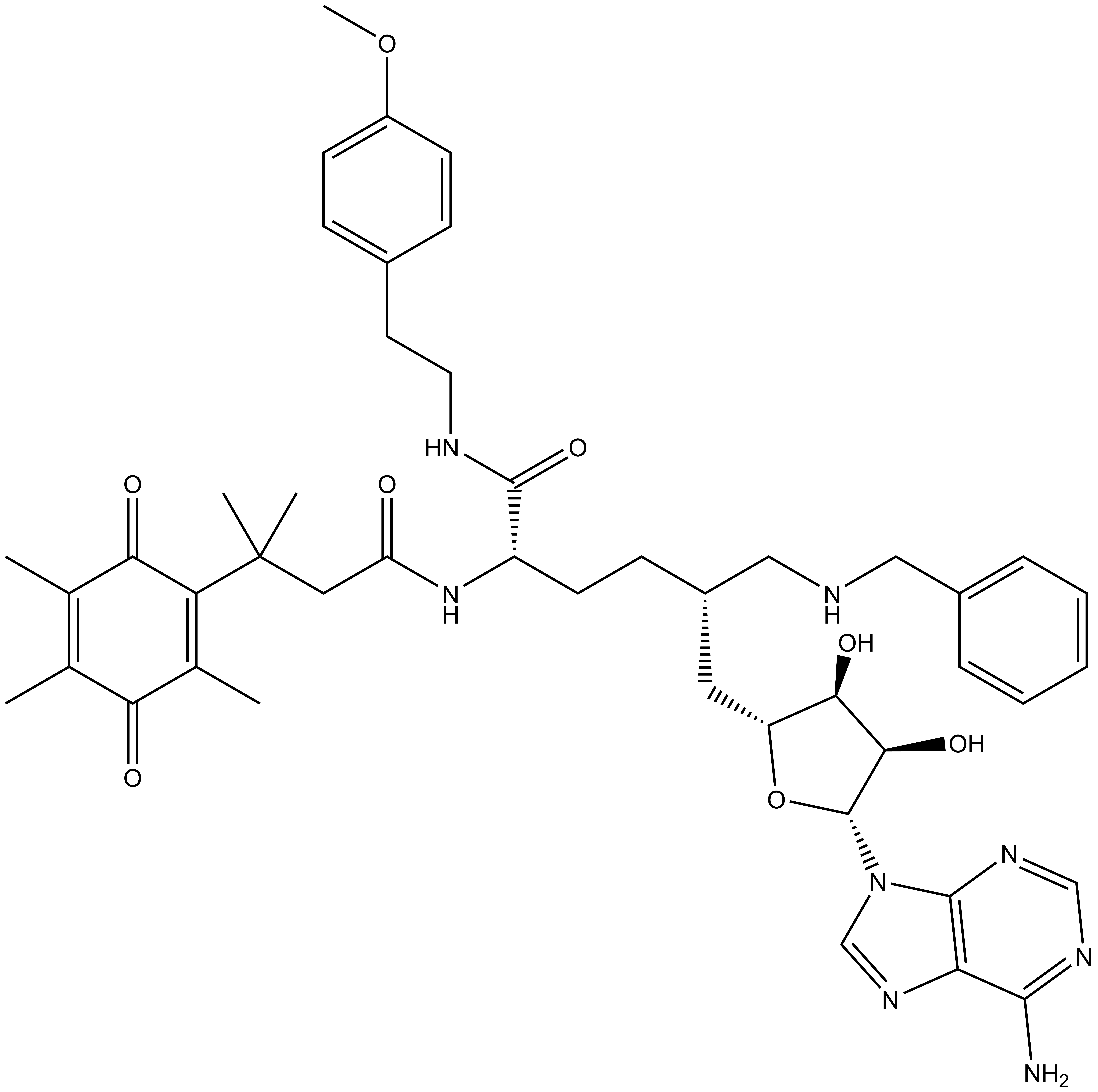
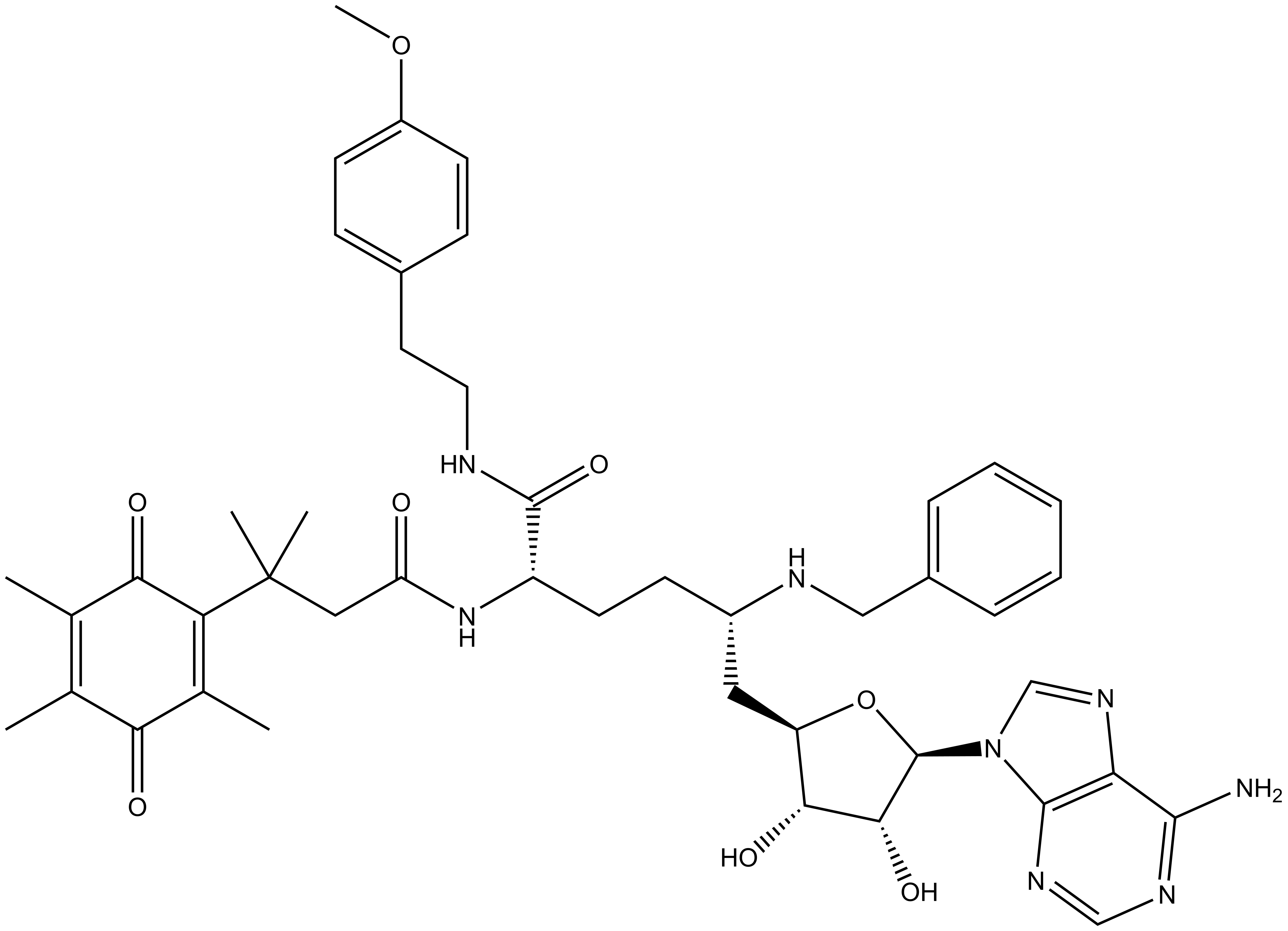
The SGC in collaboration with the Memorial Sloan Kettering Cancer Center has developed SKI-73, a chemical probe for PRMT4. SKI-73 is active in cells and is the prodrug of SKI-72, a potent and selective inhibitor of PRMT4.
Data relating to the discovery of this probe is being prepared for publication. In the meantime, in order to facilitate research by the community we are making this compound available through this website
| Probe | Negative control | |
 |
|  |
SKI-73 |
| SKI-73N |
| Physical and chemical properties for SKI-73 | |
| Molecular weight | 850.4 |
| Molecular formula | C46H58N8O8 |
| IUPAC name | 2-(3-(5-(5-(5-amino-2,4,7,9-tetraaza-bicyclo[4.3.0]nona-1(6),2,4,7-tetraen-9-yl)-3,4-dihydroxy-tetrahydro-furan-2-yl)-1-((2-(4-methoxy-phenyl)-ethylamino)-formyl)-4-((phenyl-methylamino)-methyl)-pentylamino)-1,1-dimethyl-3-oxo-propyl)-3,5,6-trimethyl-cyclohexa-2,5-diene-1,4-dione |
| MollogP | 3.724 |
| PSA | 186.5 |
| No. of chiral centres | 6 |
| No. of rotatable bonds | 21 |
| No. of hydrogen bond acceptors | 16 |
| No. of hydrogen bond donors | 7 |
| Physical and chemical properties for SKI-73N (Negative Control) | |
| Molecular weight | 836.4 |
| Molecular formula | C45H56N8O8 |
| IUPAC name | 2-(3-(5-(5-(5-amino-2,4,7,9-tetraaza-bicyclo[4.3.0]nona-1(6),2,4,7-tetraen-9-yl)-3,4-dihydroxy-tetrahydro-furan-2-yl)-1-((2-(4-methoxy-phenyl)-ethylamino)-formyl)-4-(phenyl-methylamino)-pentylamino)-1,1-dimethyl-3-oxo-propyl)-3,5,6-trimethyl-cyclohexa-2,5-diene-1,4-dione |
| MollogP | 3.239 |
| PSA | 186.2 |
| No. of chiral centres | 6 |
| No. of rotatable bonds | 20 |
| No. of hydrogen bond acceptors | 16 |
| No. of hydrogen bond donors | 7 |
Main features
This probe is available from Tocris, and Sigma
The probe and its negative control are also available from opnMe.com.
The control may be requested by clicking here.
| Probe | Negative control | |
 |
|  |
BI-9321 |
| BI-9466 |
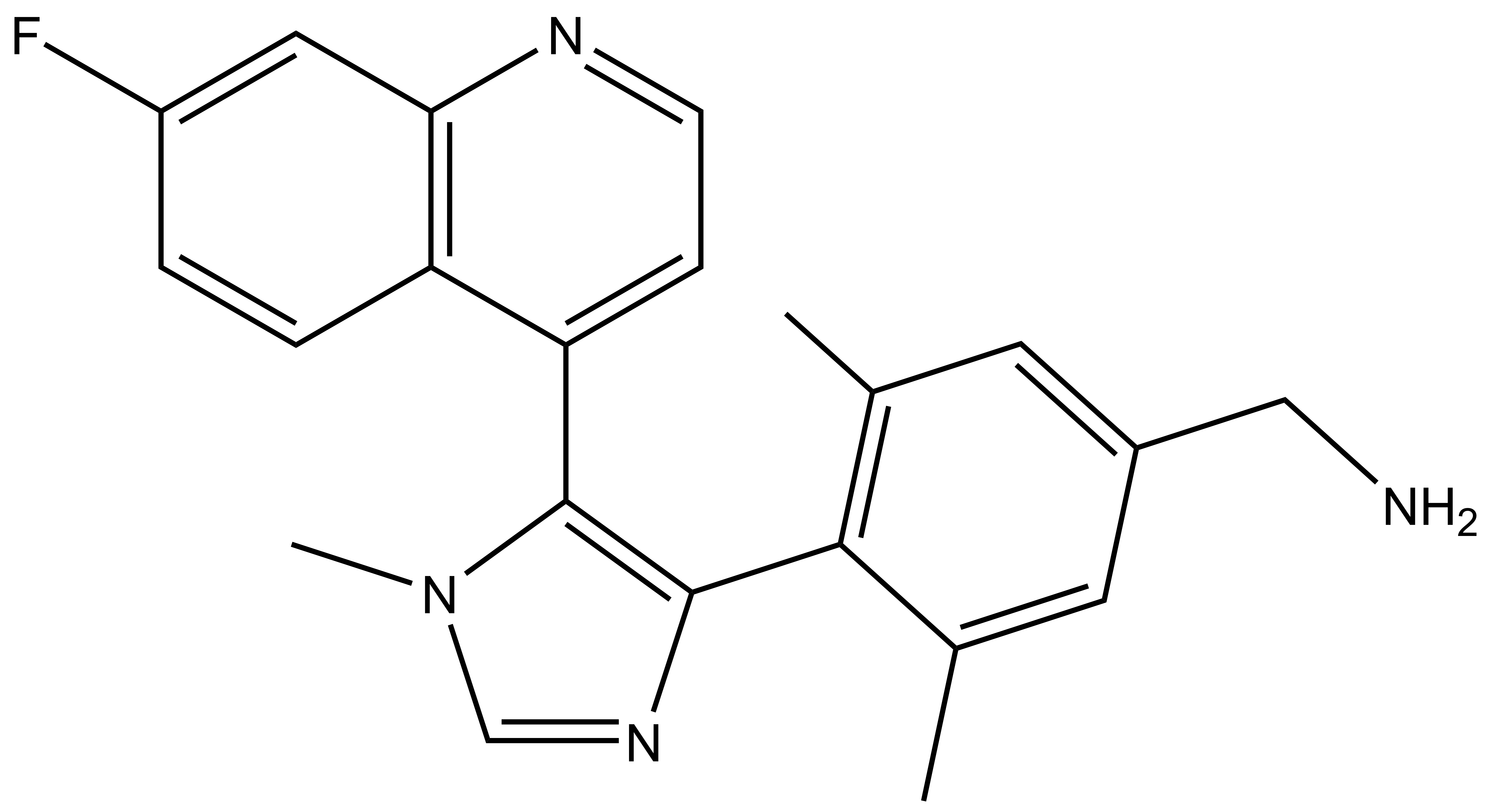
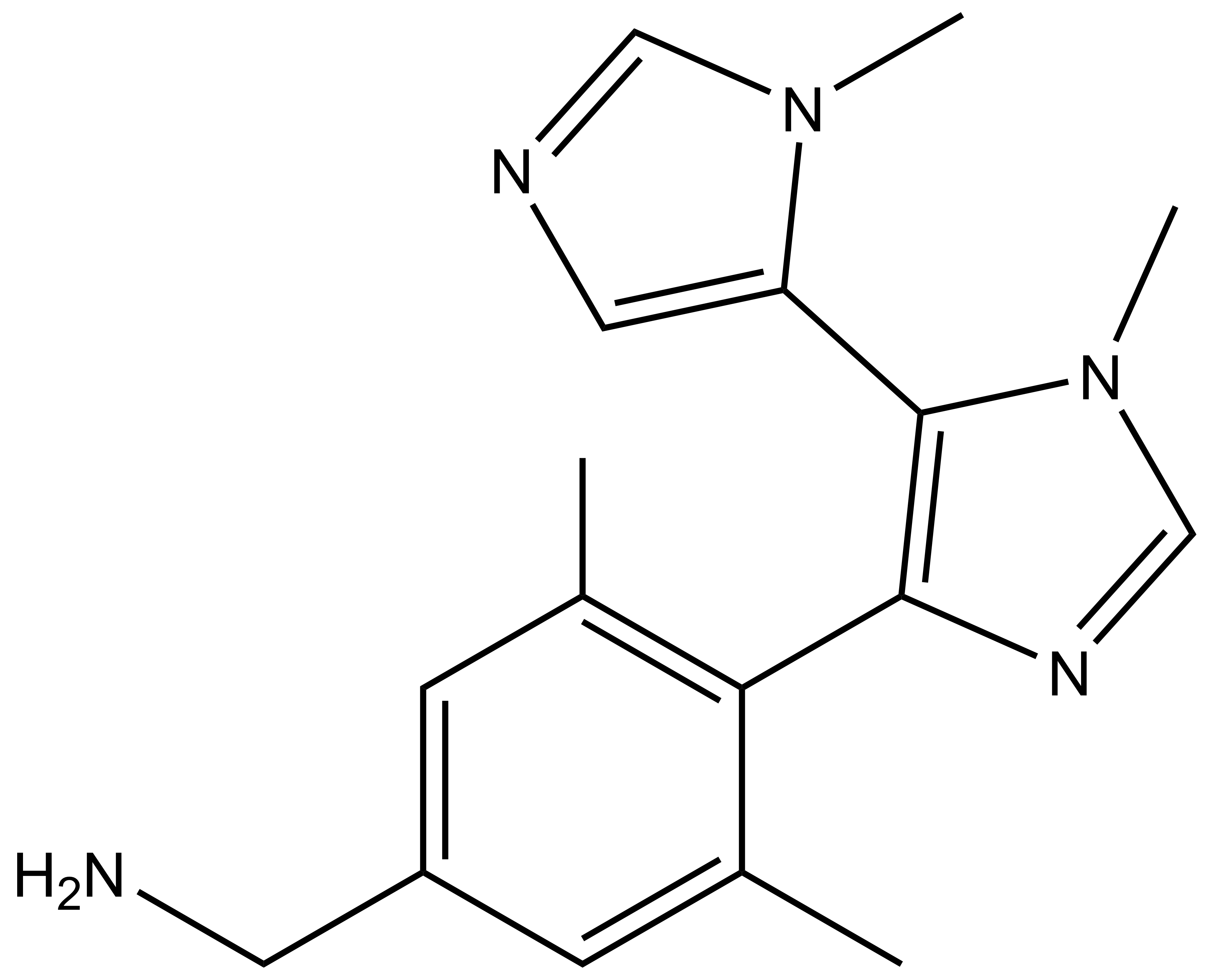
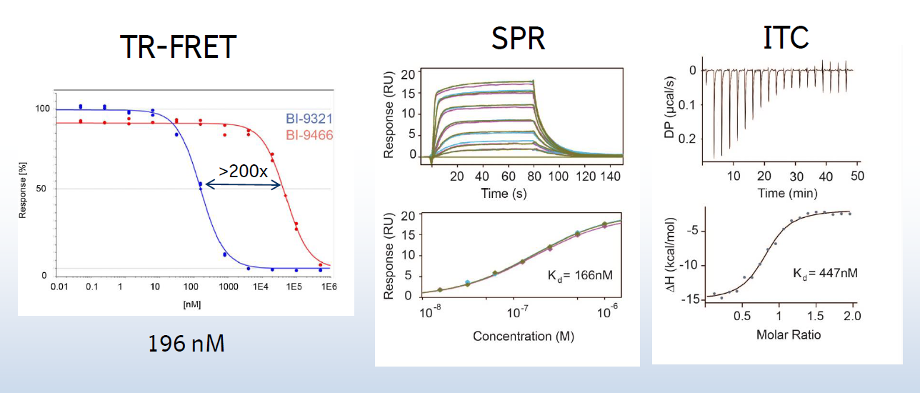
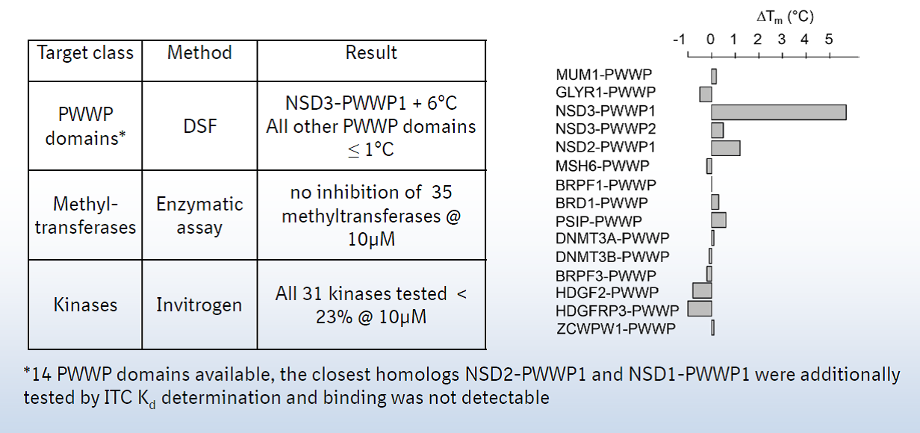
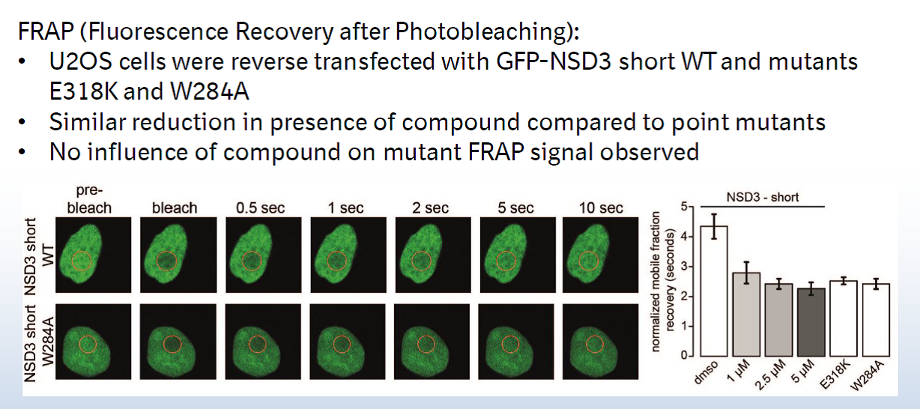
A collaboration between Boehringer Ingelheim and the SGC has resulted in the discovery of BI-9321, a potent and selective antagonist of the PWWP1 domain of NSD3. BI-9321 binds to the PWWP1 domain of NSD3 with a Kd of 166 nM by SPR and is selective over other PWWP domains including NSD2-PWWP1 and NSD3-PWWP2. BI-9321 antagonizes the interaction of H3 with NSD3-PWWP1 in U2OS cells as measured by NanoBRET with IC50 of 1.2 µM. A closely-related compound, BI-9466 is 200-fold less active and is a recommended negative control. Both compounds should be used in parallel in a dose response range between 0.1 and 20µM.
Data relating to the discovery of this probe is being prepared for publication. In the meantime, in order to facilitate research by the community we are making this compound available through this website.
| Probe | Negative control | |
 |
|  |
BI-9321 |
| BI-9466 |
| Physical and chemical properties for BI-9321 | |
| Molecular weight | 360.2 |
| Molecular formula | C22H21FN4 |
| IUPAC name | 4-(4-(amino-methyl)-2,6-dimethyl-phenyl)-5-(9-fluoro-2-aza-bicyclo[4.4.0]deca-1(6),2,4,7,9-pentaen-5-yl)-1-methyl-1H-imidazole |
| MollogP | 3.337 |
| PSA | 41.44 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 3 |
| No. of hydrogen bond acceptors | 3 |
| No. of hydrogen bond donors | 2 |
| Physical and chemical properties for BI-9466 (Negative Control) | |
| Molecular weight | 295.2 |
| Molecular formula | C17H21N5 |
| IUPAC name | 5-(5-(4-(amino-methyl)-2,6-dimethyl-phenyl)-3-methyl-3H-imidazol-4-yl)-1-methyl-1H-imidazole |
| MollogP | 0.7975 |
| PSA | 43.92 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 3 |
| No. of hydrogen bond acceptors | 3 |
| No. of hydrogen bond donors | 2 |
SMILES:
BI-9321: CC1=C(C2=C(N(C=N2)C)C3=CC=NC4=C3C=CC(F)=C4)C(C)=CC(CN)=C1
BI-9466: CC1=C(C2=C(N(C=N2)C)C3=CN=CN3C)C(C)=CC(CN)=C1
InChI:
BI-9321: InChI=1S/C22H21FN4/c1-13-8-15(11-24)9-14(2)20(13)21-22(27(3)12-26-21)18-6-7-25-19-10-16(23)4-5-17(18)19/h4-10,12H,11,24H2,1-3H3
BI-9466: InChI=1S/C17H21N5/c1-11-5-13(7-18)6-12(2)15(11)16-17(22(4)10-20-16)14-8-19-9-21(14)3/h5-6,8-10H,7,18H2,1-4H3
InChIKey:
BI-9321: WOAOENGFAAUUGT-UHFFFAOYSA-N
BI-9466: SFZHMKDAVPIXRB-UHFFFAOYSA-N
Main features
| Probe | Negative control | |
 |
|  |
MRK-740 |
| MRK-740-NC |
A collaboration between MSD and the SGC has resulted in the discovery of MRK-740, a potent inhibitor of PRDM9 with a peptide competitive MOA. MRK-740 inhibits in vitro methylation of H3K4 with IC50 = 85 nM and shows more than 100-fold selectivity over other histone methyltransferases and other non-epigenetic targets. MRK-740 inhibits the methylation of H3K4 in cells with IC50 = 0.8 µM. A control compound, MRK-740-NC, has also been developed which inhibits the in vitro methylation of H3K4 with IC50 > 100 µM. The use of this compound at 3 µM is recommended in cells.
Data relating to the discovery of this probe is being prepared for publication. In the meantime, in order to facilitate research by the community we are making this compound available through this website.
| Probe | Negative control | |
 |
|  |
MRK-740 |
| MRK-740-NC |
| Physical and chemical properties for MRK-740 | |
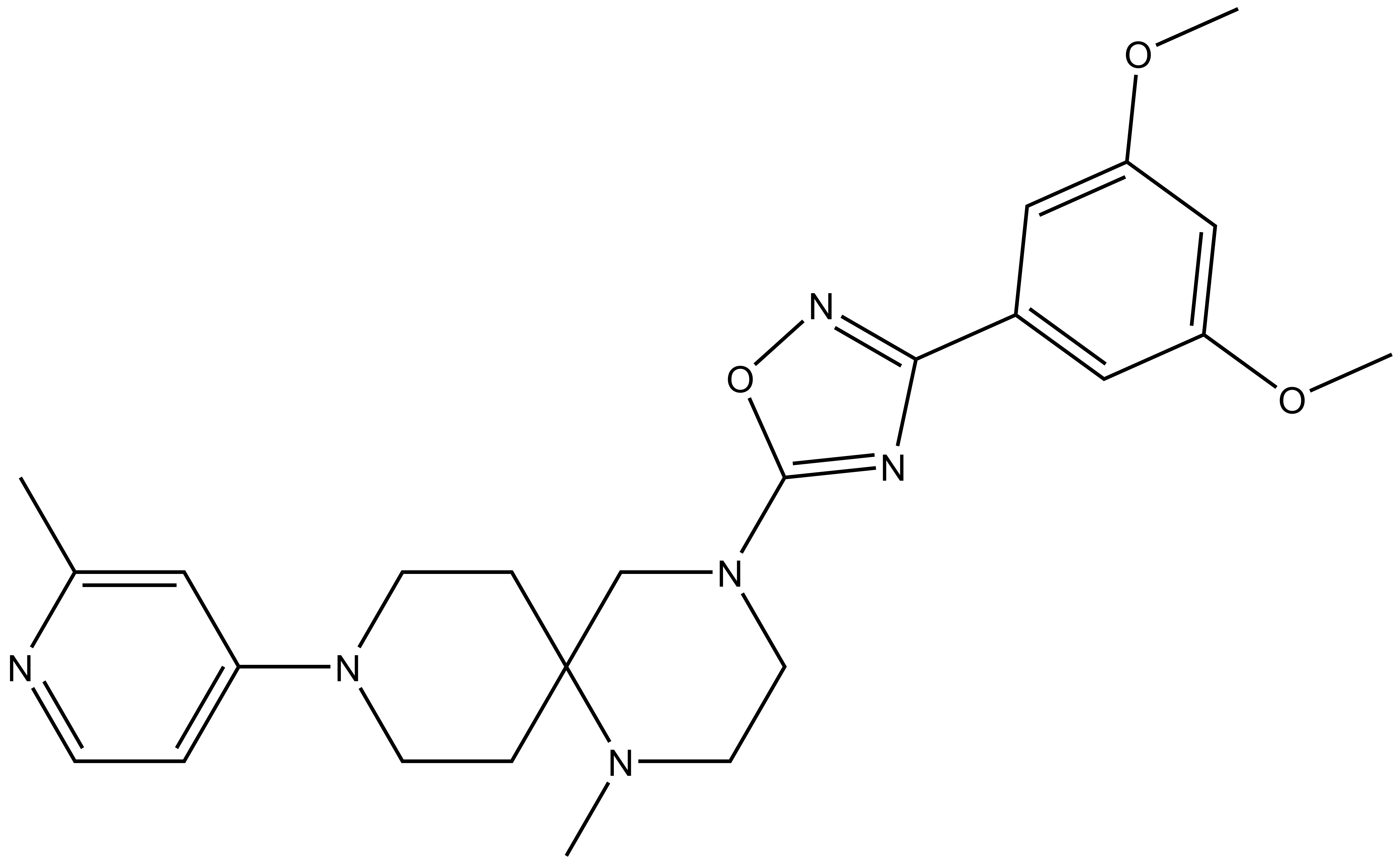
| Molecular weight | 464.3 |
| Molecular formula | C25H32N6O3 |
| IUPAC name | 4-(3-(3,5-dimethoxy-phenyl)-1,2,4-oxadiazol-5-yl)-1-methyl-9-(2-methyl-pyridin-4-yl)-1,4,9-triaza-spiro[5.5]undecane |
| MollogP | 2.851 |
| PSA | 65.67 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 7 |
| No. of hydrogen bond acceptors | 5 |
| No. of hydrogen bond donors | 0 |
| Physical and chemical properties for MRK-740-NC (Negative Control) | |
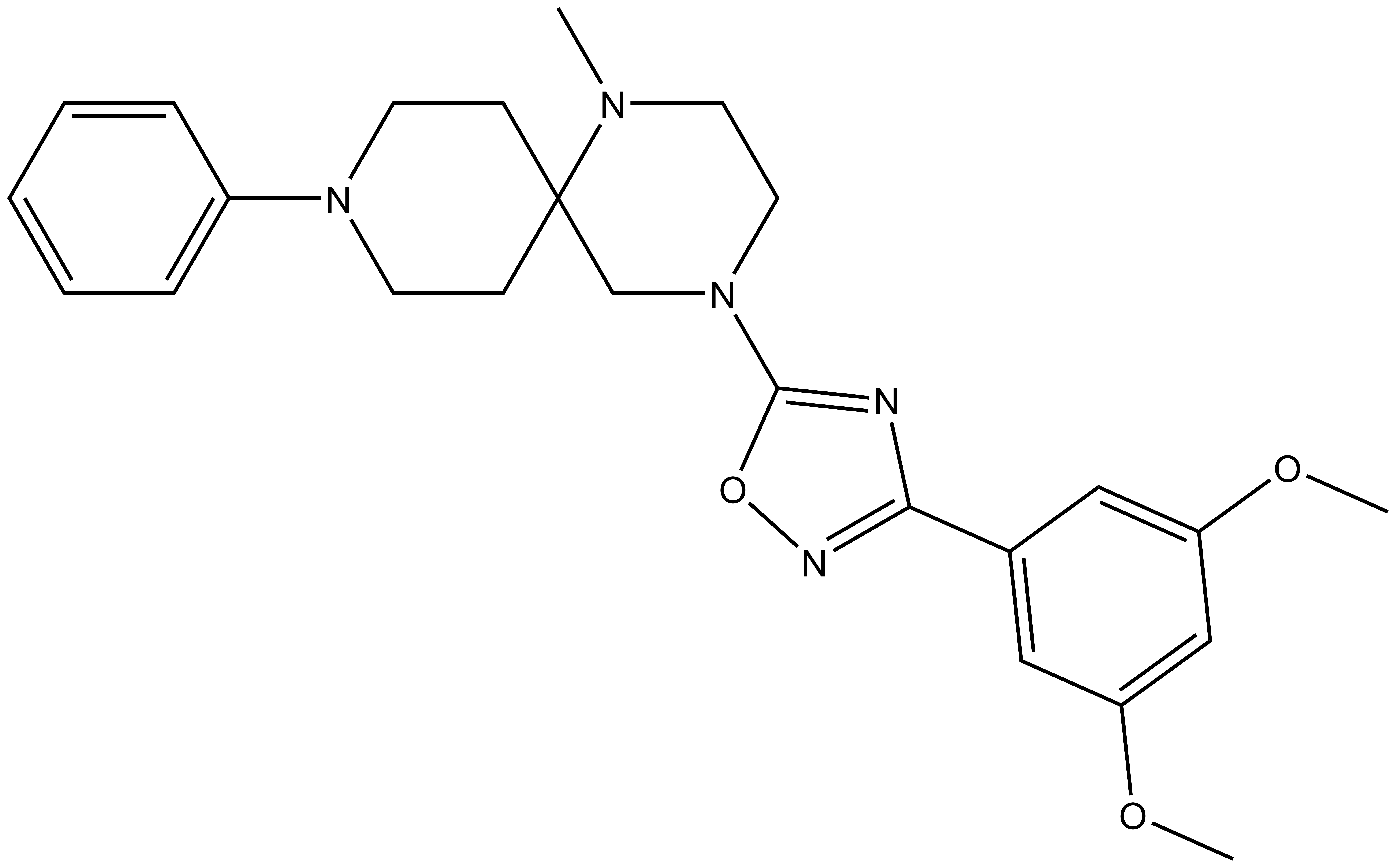
| Molecular weight | 449.2 |
| Molecular formula | C25H31N5O3 |
| IUPAC name | 4-(3-(3,5-dimethoxy-phenyl)-1,2,4-oxadiazol-5-yl)-1-methyl-9-phenyl-1,4,9-triaza-spiro[5.5]undecane |
| MollogP | 3.519 |
| PSA | 56.81 |
| No. of chiral centres | 0 |
| No. of rotatable bonds | 5 |
| No. of hydrogen bond acceptors | 6 |
| No. of hydrogen bond donors | 0 |
SMILES:
MRK-740: CC1=CC(N2CCC3(CN(C4=NC(C5=CC(OC)=CC(OC)=C5)=NO4)CCN3C)CC2)=CC=N1
MRK-740-NC: CN1CCN(C2=NC(C3=CC(OC)=CC(OC)=C3)=NO2)CC14CCN(C5=CC=CC=C5)CC4
InChI:
MRK-740: InChI=1S/C25H32N6O3/c1-18-13-20(5-8-26-18)30-9-6-25(7-10-30)17-31(12-11-29(25)2)24-27-23(28-34-24)19-14-21(32-3)16-22(15-19)33-4/h5,8,13-16H,6-7,9-12,17H2,1-4H3
MRK-740-NC: InChI=1S/C25H31N5O3/c1-28-13-14-30(18-25(28)9-11-29(12-10-25)20-7-5-4-6-8-20)24-26-23(27-33-24)19-15-21(31-2)17-22(16-19)32-3/h4-8,15-17H,9-14,18H2,1-3H3
InChIKey:
MRK-740: NZYTZRHHBAJPKN-UHFFFAOYSA-N
MRK-740-NC: OACWMVMQWRVMAF-UHFFFAOYSA-N
Main features
With this unique project, several pharmaceutical companies (AbbVie, Bayer, Boehringer Ingelheim, Janssen, MSD, Pfizer, and Takeda) have entered into a pre-competitive collaboration with the SGC, to make a large number of innovative high-quality probes available to the research community which can be found here: http://www.sgc-ffm.uni-frankfurt.de/.