SGC chemical probes are open-access reagents that are meant to be used by the biomedical research community with no restrictions on use. SGC-developed probes are complemented by a growing set of donated high-quality chemical probes developed in industry and non-SGC laboratories. These are also available for unencumbered use.
The table below contains the entire collection of probes from SGC’s chemical probes program and donated chemical probes. To browse the collection, find associated data, or acquire samples, select the desired chemical probe in the table below and follow the links that appear on that page. You can sort the table by clicking the column headers.
For inquiries or requests related to chemical probes or libraries, please contact us at proberequests@thesgc.org.
| Name | Structures | Protein Family | Target Protein | Negative Control | Cellular usage | Added |
|---|---|---|---|---|---|---|
| JNJ-8003 |
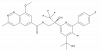
|
RNA polymerase | RSV-L | JNJ-6643 | ||
| JNJ-78911118 |
|
Glutamate-gated ion channel | GRIN2A | JNJ-77914993 | up to 10 µM | |
| ACBI3 |
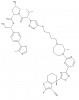
|
GTPase | KRAS | cis-ACBI3 | 1-10 µM | |
| THNAN69 |
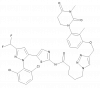
|
Kinase | LIMK2 | THNAN69-NC | 10 -100 nM | |
| SM311 |
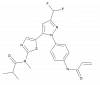
|
Kinase | LIMK1 | SM311-NC | 0.1 - 1 µM | |
| BI-5121 |
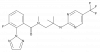
|
GPCR | HCRTR1 | BI-6199 | 100 nM | |
| BAY-439 |
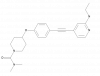
|
Phospholipase A2 | PLA2G5 | BAY-163 | up to 300 nM | |
| SGC-NSP2hel-1 |
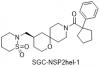
|
Helicases | CHIKV_nsp2 helicase | SGC-NSP2hel-1N | 1 µM | |
| JYQ-173 |
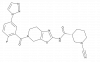
|
Peptidase C56 | PARK7 | MB078 | Up to 1 µM | |
| Huib32 |
|
Deubiquitinase | USP32 | Huib32NC | 5 µM | |
| JNJ-79883960 |
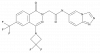
|
Receptor | NRLP3 | JNJ-78832000 | up to 10 µM | |
| SGC-NSP2PRO-1 |
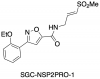
|
Protease | CHIKV_nsp2 | SGC-NSP2PRO-1N | ||
| LH168 |
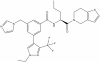
|
WD40 repeat | WDR5 | LH222 | 1 µM | |
| OICR-41103 |
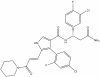
|
WD40 repeat | DCAF1 | OICR-41103N | 1 µM; not to exceed 5 µM | |
| BI-1915 |

|
Protease | CTSS | BI-1920 | 1 µM | |
| BI-113823 |
|
GPCR | BDKRB1 | BI-5832 | 100 nM | |
| JNJ-54082730 |
|
Hydrolase | PDE2A | JNJ-54103985 | 1 µM | |
| JNJ-3738 |
|
Kinase | CDK7 | JNJ-6240 | 1 µM | |
| BAY-9835 |
|
Protease | ADAMTS7, ADAMTS12 | BAY-1880 | 100 nM | |
| SGC-CDKL2/AAK1/BMP2K-1 |
|
Kinase | CDKL2 | CDKL2/AAK1/BMP2K -1N | ≤1 µM | |
| ABT-724 |
|
GPCR | DRD4 | A-769 | ||
| ERKi |

|
Kinase | MAPK3, MAPK1 | ERKi-NC | ||
| BAY-7598 |

|
Protease | MMP12 | BAY-694 | ||
| BI 99179 |
|
Synthase | FASN | BI 99990 | ||
| BAY-474 |

|
Kinase | MET | BAY-827 | ||
| (R)-9s |
|
GPCR | ADRA1D | (S)-9s | ||
| ABT-100 |

|
Transferase | FNTB | A-108 | ||
| BTZO-1 |

|
Cytokine | MIF | BTZO-4 | ||
| BAY-386 |

|
GPCR | F2R | BAY-448 | ||
| BAY-707 |

|
Hydrolase | NUDT1 | BAY-604 | ||
| A-485 |
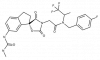
|
Transferase | EP300, CREBBP | A-486 | ||
| BI-9627 |
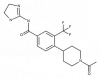
|
Solute carriers | SLC9A1 | BI-0054 | ||
| TP-004 |
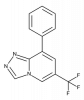
|
Protease | METAP2 | TPn-004 | ||
| PF-05105679 |
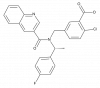
|
Ion channel | TRPM8 | PF-05257137 | ||
| PF-04418948 |
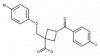
|
GPCR | PTGER2 | PF-04475866 | ||
| BI-1935 |
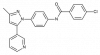
|
Hydrolase | EPHX2 | BI-2049 | ||
| GSM1 |
|
Protease | Gamma secretase complex | GSM-NC | ||
| MRK-560 |

|
Protease | Gamma secretase complex | GSI-NC | ||
| MRL-SYKi |
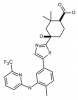
|
Kinase | SYK | MRL-SYKi-NC | ||
| MRL-650 |

|
GPCR | CNR1 | MRL-CB1-NC | ||
| CRTH2 antagonist |
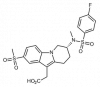
|
GPCR | PTGDR2 | CRTH2 negative control | ||
| TP-021 |
|
Transcription factor | BCL6 | TP-021n | ||
| TP-020 |
|
Transferase | MGAT2 | TP-020n | ||
| BAY-784 |
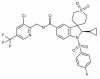
|
GPCR | GNRHR | BAY-786 | ||
| BAY-1797 |
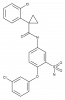
|
Ion channel | P2X4 | BAY-207 | ||
| BI-1950 |
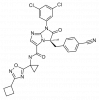
|
Integrin | ITGAL | BI-9446 | ||
| BI 653048 |
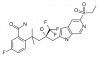
|
Nuclear hormone receptor | NR3C1 | BI-3047 | ||
| BI 639667 |
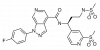
|
GPCR | CCR1 | BI-9307 | ||
| BI-2545 |
|
Phosphodiesterase | ENPP2 | BI-3017 | ||
| BAY-885 |
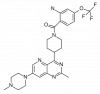
|
Kinase | MAPK7 | BAY-693 | ||
| BI-4394 |
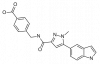
|
Protease | MMP13 | BI-4395 | ||
| A-079 |
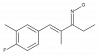
|
Ion channel | TRPA1 | A-226 | ||
| BAY-678 |
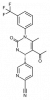
|
Protease | ELANE | BAY-677 | ||
| BAY-876 |
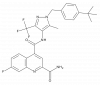
|
Solute carriers | SLC2A1 | BAY-588 | ||
| T-26c |
|
Protease | MMP13 | T-26f | ||
| ABT-546 |

|
GPCR | EDNRA | A-545 | ||
| PF-04457845 |

|
Hydrolase | FAAH | PF-04875474 | ||
| KISS1-305 |
|
GPCR | KISS1R | KISS1-543 | ||
| A-1155463 |
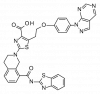
|
Regulator protein | BCL2L1 | A-1107969 | ||
| A-1211212 |
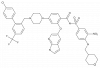
|
Regulator protein | BCL2 | A-1210227 | ||
| IPP/CNRS-A017 |
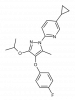
|
Dehydrogenase | DHODH | IPP/CNRS-A019 | ||
| MSD-M1PAM |

|
GPCR | CHRM1 | MSD-M1PAM-NC | ||
| A-1596586 |

|
Ion channel | CFTR | A-1596584 | ||
| MSD-CYP11B2 |
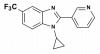
|
Cytochrome P450 | CYP11B2 | MSD-CYP11B2 Negative control | ||
| THPP-1 |
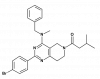
|
Phosphodiesterase | PDE10A | THPP-1-NC | ||
| PPTN |
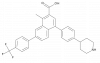
|
GPCR | P2RY14 | PPTN-NC | ||
| BAY-549 |
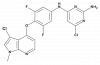
|
Kinase | ROCK1, ROCK2 | BAY-4900 | ||
| TP-008 |
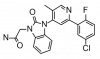
|
Kinase | ACVR1B, TGFBR1 | Al11 | ||
| MLi-2 |
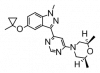
|
Kinase | LRRK2 | MLi-2-NC | ||
| UCSF924 |
|
GPCR | DRD4 | UCSF924NC | ||
| T3-CLK |
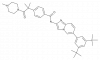
|
Kinase | CLK1, CLK2, CLK3, CLK4 | T3-CLK-N | ||
| BAY-293 |
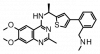
|
Nucleotide exchange factor | SOS1 | BAY-294 | ||
| A-192621 |
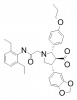
|
GPCR | EDNRB | A-1806262 | ||
| BI-1230 |
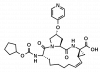
|
Protease | HCV NS3 | BI-1675 | ||
| TP-024 |
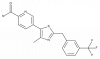
|
GPCR | GPR52 | TP-024n | ||
| BI-2540 |
|
Reverse transcriptase | HIV NNRT | BI-2439 | ||
| BAY-390 |
|
Ion channel | TRPA1 | BAY-9897 | ||
| BAY-069 |
|
Amino acid transaminase | BCAT1, BCAT2 | BAY-771 | ||
| TP-030-2 |
|
Kinase | RIPK1 | TP-030n | ||
| TP-030-1 |
|
Kinase | RIPK1 | TP-030n | ||
| (R)-ZINC-3573 |
|
GPCR | MRGPRX2 | (S)-ZINC-3573 | ||
| Ogerin |
|
GPCR | GPR68 | ZINC32547799 | ||
| WM-1119 |
|
Transferase | KAT6A, KAT6B | WM-2474 | ||
| SR-302 |
|
Kinase | DDR1, DDR2, MAPK11, MAPK14 | SR-301 | ||
| NVS-MALT1 |
|
Protease | MALT1 | NVS-MALT1-C | ||
| BAY-3827 |
|
Kinase | PRKAA1, RPS6KA1 | BAY-974 | ||
| FS-694 |
|
Kinase | MAPK14 | FM-743 | ||
| Skepinone-L |
|
Kinase | MAPK14 | FM-743 | ||
| SR-318 |
|
Kinase | MAPK14 | SR-321 | ||
| BAY-179 |
|
Protease | Complex I | BAY-070 | Yes | |
| BAY-899 |
|
GPCR | LHCGR | BAY-897 | ||
| BAY-985 |
|
Kinase | TBK1, IKBKE | BAY-440 | ||
| Borussertib |
|
Kinase | AKT1, AKT2 | RL2578 | ||
| BAY-091 |
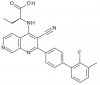
|
Kinase | PIP4K2A | BAY-0361 | ||
| MSC-4381 |
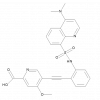
|
Monocarboxylate transporter | SLC16A3 | MSC-0516 | ||
| PFI-653 |
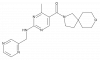
|
Hydrolase | VNN1 | PFI-653-N | ||
| JNJ-54119936 |
|
Nuclear hormone receptor | RORC | JNJ-53721590 | ||
| JNJ-42396302 | Phosphodiesterase | PDE10A | JNJ-40573663 | |||
| TP-040 |
|
Hydrolase | OGA | TP-040n | ||
| 8RK64 |
|
Regulator protein | UCHL1 | JYQ88 | ||
| A-446 |
|
Hydrolase | GLS | A-426 | ||
| BI-1942 | Protease | CMA1 | ||||
| BI 605906 |
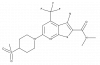
|
Kinase | IKBKB | BI-5026 | ||
| GSK973 |
|
Bromodomains | BRD2, BRD3, BRD4, BRDT | GSK943 | ||
| GSK620 | Bromodomains | BRD2, BRD3, BRD4, BRDT | ||||
| GSK046 | Bromodomains | BRD2, BRD3, BRD4, BRDT | ||||
| GSK789 |
|
Bromodomains | BRD2, BRD3, BRD4, BRDT | GSK791 | ||
| GSK778 | Bromodomains | BRD2, BRD3, BRD4, BRDT | ||||
| BAY-3153 |
|
GPCR | CCR1 | BAY-173 | ||
| BAY-6672 |
|
GPCR | PTGFR | BAY-403 | ||
| BI 207127 |
|
RNA polymerase | HCV NS5B | BI-7656 | ||
| BAY 1816032 |
|
Kinase | BUB1 | BAY-283 | ||
| JNJ-9350 |
|
Oxidase | SMOX | JNJ-4545 | ||
| SAFit2 |
|
Isomerase | FKBP5 | ddSAFit2 | ||
| CCT369260 |
|
Transcription factor | BCL6 | CCT393732 | ||
| JNJ-39758979 |
|
GPCR | HRH4 | JNJ-39668551 | ||
| JNJ-6204 |
|
Kinase | CSNK1D/CSNK1E | JNJ-0293 | ||
| BI-2081 |
|
GPCR | FFAR1 | BI-0340 | ||
| TP-051 |
|
GPCR | FFAR1 | TP-051n | ||
| TP-050 |
|
Ion channel | GRIN2A | TP-050n | ||
| GNE-2256 |
|
Kinase | IRAK4 | GNE-6689 | ||
| BAY1125976 |
|
Kinase | AKT1/2 | BAY-940 | ||
| JNJ-65355394 |
|
Hydrolase | OGA | JNJ-73924149 | ||
| TP-061 |
|
Helicases | Brr2 | TP-061N | 5 µM | |
| SGC-UBD1031 |
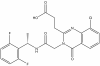
|
UBD (ubiquitin binding domain) | USP16/HDAC6 | SGC-UBD1031N | 10 µM | |
| BAY 1816032 |

|
2OG | BUB1 | BAY-283 | ||
| UNC8732 |
|
Methyl Lysine Binder | NSD2 | UNC8884 | 3 µM | |
| OICR11029 |
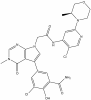
|
Transcription factor | BCL6 | OICR11600 | 1 µM | |
| PFI-8 |
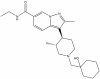
|
YEATS | YEATS | PFI-8N | 1 µM | |
| BI-5524 |
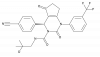
|
Chymotrypsin | ELANE | BI-5525 | ||
| WEB2086 |
|
GPCR | PTAFR | WEB2387 | ||
| BIBO3304 |
|
GPCR | NPY1R | BIBO3457 | ||
| SGK3-PROTAC1 |
|
Kinase | SGK3 | cisSGK3-PROTAC1 | ||
| VZ185 |
|
Bromodomain-containing protein family | BRD7, BRD9 | cisVZ185 | ||
| BAY-277 |
|
Aminopeptidase | METAP2 | BAY-8805 | ||
| BAY-6096 |
|
GPCR | ADRA2B | BAY-726 | ||
| BAY 1753011 |
|
GPCR | AVPR1A, AVPR2 | BAY-2297 | ||
| JA310 |
|
Kinase | MST3, MST4 | JA262 | < 1 µM | |
| BI-8668 |
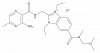
|
Sodium channels | Epithelial sodium channel (ENaC) | BI-0337 | ||
| BI-3812 |
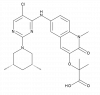
|
Transcription factor | BCL6 | BI-5273 | ||
| BI-3802 |
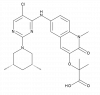
|
Transcription factor | BCL6 | BI-5273 | ||
| JP3000 |
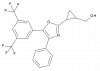
|
Nuclear hormone receptor | RXRA, RXRB, RXRG | JP3001 | ||
| SGC-GSK3-1 |
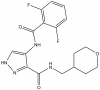
|
Kinase | GSK3 | SGC-CDKL5/GSK3-1N | 100 nM | |
| SGC-CDKL5/GSK3 |
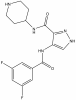
|
Kinase | CDKL5, GSK3 | SGC-CDKL5/GSK3-1N | 100 nM | |
| SGC-CAF382-1 |
|
Kinase | CDKL5 | SGC-CAF268-1N | ≤ 100 nM | |
| TP-060 |
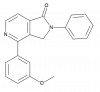
|
Glucosyltransferase | UGCG | TP-060n | ||
| JNJ-42226314 |
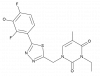
|
Serine hydrolase | MGLL | JNJ-8034 | ||
| JNJ-4355 |
|
BCL2 protein family | MCL1 | JNJ-78732576 | ||
| BAY-7081 |

|
Phoshodiesterase | PDE9A | BAY-7424 | ||
| FHT-2344 |
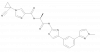
|
SWI/SNF | SMARCA2, SMARCA4 | FHT-5908 | ||
| BI-3231 |
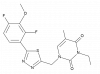
|
17β-hydroxysteroid dehydrogenases | HSD17B13 | BI-0955 | ||
| TH-257 |
|
Kinase | LIMK1, LIMK2 | TH-263 | 1 µM | |
| JA397 |
|
Kinase | CDK14-18 | JA314 | < 1 µM | |
| SGC-PI5P4Kγ/MYLK4-1 |
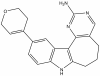
|
Kinase | PI5P4K gamma, MYLK4 | SGC-PI5P4Kγ/MYLK4-1N | 100 nM | |
| SGC-CK2-2 |
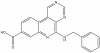
|
Kinase | CK2/CSNK2 | SGK-CK2-2N | 1 µM | |
| LIJTF500025 |
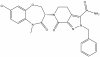
|
Kinase | LIMK1, LIMK2 | LIJTF500120 | < 1 µM | |
| MSC1186 |
|
Kinase | SRPK | MSC5360 | < 1 µM | |
| CS640 |
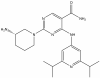
|
Kinase | CAMK1 | CS640s | < 1 µM | |
| MU1742 |
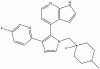
|
Kinase | CK1delta, CK1epsilon | MU2027 | 1 µM | |
| BAY-805 |
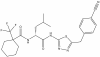
|
USP (ubiquitin specific protease) | USP21 | BAY-728 | 1 µM | |
| MRK-952 |
|
Hydrolase | NUDT5 | MRK-952-NC | < 1 µM | |
| MRK-990 |
|
Methyltransferase | PRMT9, PRMT5 | MRK-990-NC | < 10 µM | |
| SGC-PIKFYVE-1 |
|
Kinase | PIKfyve | SGC-PIKFYVE-1N | 100 nM | |
| MU1700 |
|
Kinase | ALK1, ALK2 | MU1700NC | 1 µM | |
| M4K2234 |
|
Kinase | ALK1, ALK2 | M4K2234NC | 1 µM | |
| SGC-UBD253 |
|
UBD (ubiquitin binding domain) | HDAC6 | SGC-UBD253N | 3 µM | |
| CK156 |
|
Kinase | STK17A, DRAK1 | CKJB71 | 5 µM | |
| PFI-7 |
|
E3 ligase | GID4 | PFI-7N | 1 µM | |
| MRIA9 |
|
Kinase | SIK1, SIK2, SIK3 | MR7 | 10 µM | |
| Homer |
|
WD40 repeat | WDR5 | nc_WDR5, nc_VHL | 1 µM | |
| SGC-SMARCA-BRDVIII |
|
Bromodomains | PB1(5), SMARCA2/4 | SGC-BRDVIII-NC | 10 µM | |
| NR162 |
|
Kinase | CASK | NR187 | 10 µM | |
| PFI-6 |
|
YEATS | MLLT1, MLLT3 | PFI-6N | 1 µM | |
| SGC-CK2-1 |
|
Kinase | CSNK2A1, CSNK2A2 | SGC-CK2-1N | 100 nM | |
| NVS-MLLT-1 |
|
YEATS | MLLT1, MLLT3 | NVS-MLLT-C | 1 µM | |
| SGC-STK17B-1 |
|
Kinase | STK17B, DRAK2 | SGC-STK17B-1N | 1 µM | |
| SGC-CLK-1 |
|
Kinase | CLK1, CLK2, CLK4 | SGC-CLK-1N | 1 µM | |
| SGC-CAMKK2-1 |
|
Kinase | CAMKK1, CAMKK2 | SGC-CAMKK2-1N | 1 µM | |
| PFI-5 |
|
Methyltransferase | SMYD2 | PFI-5N | 1 µM | |
| SGC6870 |
|
Methyltransferase | PRMT6 | SGC6870N | 1 µM | |
| UNC6934 |
|
Methyl Lysine Binder | NSD2-PWWP1 | UNC7145 | 1 µM | |
| MU1210 |
|
Kinase | CLK1, CLK2, CLK4, DYRK1A, DYRK1B, DYRK2 | MU140 | 1 µM | |
| NVS-BPTF-1 |
|
Bromodomains | BPTF | NVS-BPTF-C | < 1 µM | |
| VinSpinIn |
|
Spindlin domains | Spin1 | VinSpinIC | 0.5 µM - 3 µM | |
| TP-238 |
|
Bromodomains | CECR2, BPTF | TP-422 | < 2 µM | |
| DDR-TRK-1 |
|
Kinase | DDR, TRK | DDR-TRK-1N | 5 µM | |
| SKI-73 |
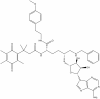
|
Methyltransferase | CARM1 | SKI-73N | 1 µM | |
| BI-9321 |
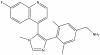
|
Methyl Lysine Binder | NSD3 | BI-9466 | 0.5 µM - 20 µM | |
| MRK-740 |
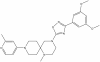
|
Methyltransferase | PRDM9 | MRK-740-NC | 3 µM | |
| BAY-826 |

|
Kinase | TIE1, TEK, DDR1, DDR2 | BAY-309 | ||
| BI-665915 |
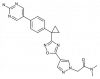
|
Lipoxygenase | ALOX5AP | BI-0153 | ||
| SGC3027 |
|
Methyltransferase | PRMT7 | SGC3027N | 1 µM | |
| SGC-GAK-1 |
|
Kinase | GAK | SGC-GAK-1N | 1 µM | |
| SGC-AAK1-1 |
|
Kinase | AAK1, BMP2K, BIKE | SGC-AAK1-1N | 1 µM | |
| BI01383298 |
|
Solute carriers | SLC13A5 | BI01372674 | 1 µM | |
| L-Moses |
|
Bromodomains | KAT2A, KAT2B | D-Moses | 1 µM | |
| BAY-6035 |
|
Methyltransferase | SMYD3 | BAY-444 | 1 µM | |
| GSK4027 |
|
Bromodomains | KAT2A, KAT2B | GSK4028 | 1 µM | |
| FM-381 |
|
Kinase | JAK3 | FM-479 | 100 nM | |
| BAY-850 |
|
Bromodomains | ATAD2 | BAY-460 | 1 µM | |
| GSK8814 |
|
Bromodomains | ATAD2, ATAD2B | GSK8815 | 1 µM | |
| GSK6853 |
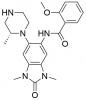
|
Bromodomains | BRPF1 | GSK9311 | < 1 µM | |
| LLY-283 |
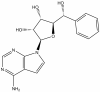
|
Methyltransferase | PRMT5 | LLY-284 | 1 µM | |
| A-395 |

|
WD40 repeat | EED | A-395N | 100 nM | |
| NVS-PAK1-1 |
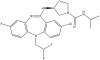
|
Kinase | PAK1 | NVS-PAK1-C | 1 µM | |
| TP-472 |
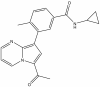
|
Bromodomains | BRD9, BRD7 | TP-472N | 1 µM | |
| TP-064 |
|
Methyltransferase | CARM1 | TP-064N | 1 µM | |
| BAY-299 |
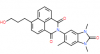
|
Bromodomains | TAF1, BRD1 | BAY-364 | 1 µM | |
| GSK864 |
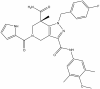
|
Dehydrogenase | IDH1 | GSK990 | 1 µM | |
| MS023 |
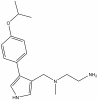
|
Methyltransferase | PRMT1, PRMT3, CARM1, PRMT6, PRMT8 | MS094 | 1 µM | |
| A-196 |
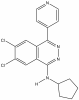
|
Methyltransferase | SUV420H1, SUV420H2 | SGC2043 | 1 µM | |
| GSK591 |
|
Methyltransferase | PRMT5 | SGC2096 | 1 µM | |
| MS049 |
|
Methyltransferase | CARM1, PRMT6 | MS049N | 1 µM | |
| NVS-CECR2-1 |
|
Bromodomains | CECR2 | NVS-CECR2-C | 1 µM | |
| I-BRD9 |
|
Bromodomains | BRD9 | 1 µM | ||
| BI-9564 |
|
Bromodomains | BRD9, BRD7 | 1 µM | ||
| BAY-598 |
|
Methyltransferase | SMYD2 | BAY-369 | 1 µM | |
| GSK484 |
|
Arginine deiminases | PAD4 | GSK106 | 1 µM | |
| GSK2801 |
|
Bromodomains | BAZ2A, BAZ2B | GSK8573 | 1 µM | |
| LP99 |
|
Bromodomains | BRD9, BRD7 | 2S,3R-enantiomer | 1 µM | |
| NI-57 |
|
Bromodomains | BRPF1, BRD1, BRPF3 | 1 µM | ||
| PFI-4 |
|
Bromodomains | BRPF1 | 1 µM | ||
| OF-1 |
|
Bromodomains | BRPF1, BRD1, BRPF3 | <5 µM | ||
| SGC707 |
|
Methyltransferase | PRMT3 | XY1 | 1 µM | |
| BAZ2-ICR |
|
Bromodomains | BAZ2A, BAZ2B | 1 µM | ||
| OICR-9429 |

|
WD40 repeat | WDR5 | OICR-0547 | 1 µM | |
| GSK-LSD1 |
|
Lysine Demethylase | KDM1A/LSD1 | 100 nM | ||
| LLY-507 (multiple off-targets) |
|
Methyltransferase | SMYD2 | SGC705 | 1 µM | |
| PFI-3 |
|
Bromodomains | SMARCA2, PBRM1, SMARCA4 | PFI-3oMet | 1 µM | |
| A-366 |
|
Methyltransferase | EHMT1/GLP, EHMT2/G9a | 1 µM | ||
| UNC1999 |
|
Methyltransferase | EZH2 | UNC2400 | 1 µM | |
| (R)-PFI-2 |
|
Methyltransferase | SETD7 | (S)-PFI-2 | 1 µM | |
| SGC-CBP30 |
|
Bromodomains | CREBBP, EP300 | BDOIA513 | 1 µM | |
| I-CBP112 |
|
Bromodomains | CREBBP, EP300 | 1 µM | ||
| Bromosporine (BSP) |
|
Bromodomains | pan-Bromodomain | 5 µM | ||
| UNC1215 |
|
Methyl Lysine Binder | L3MBTL3 | UNC1079 | 1 µM | |
| UNC0642 |
|
Methyltransferase | EHMT2/G9a, EHMT1/GLP | 1 µM | ||
| GSK343 |
|
Methyltransferase | EZH2 | 1 µM | ||
| GSK-J1 |
|
Lysine Demethylase | KDM6B/JMJD3, KDM6A/UTX, KDM5B/JARID1B | GSK-J2 (GSK-J5) | 100 nM | |
| SGC0946 |
|
Methyltransferase | DOT1L | SGC0649 | 1 µM | |
| IOX2 |
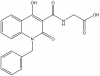
|
2OG | PHD2 | 1 µM | ||
| IOX1 |
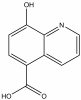
|
2OG | Pan-KDM | 1 µM - 20 µM | ||
| PFI-1 |
|
Bromodomains | BRD2, BRD3, BRD4, BRDT | 1 µM | ||
| UNC0638 |
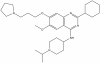
|
Methyltransferase | EHMT2/G9a, EHMT1/GLP | UNC0737 | ≤ 1 µM | |
| (+)-JQ1 |
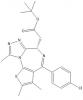
|
Bromodomains | BRD2, BRD3, BRD4, BRDT | (-)-JQ1 | 1 µM |